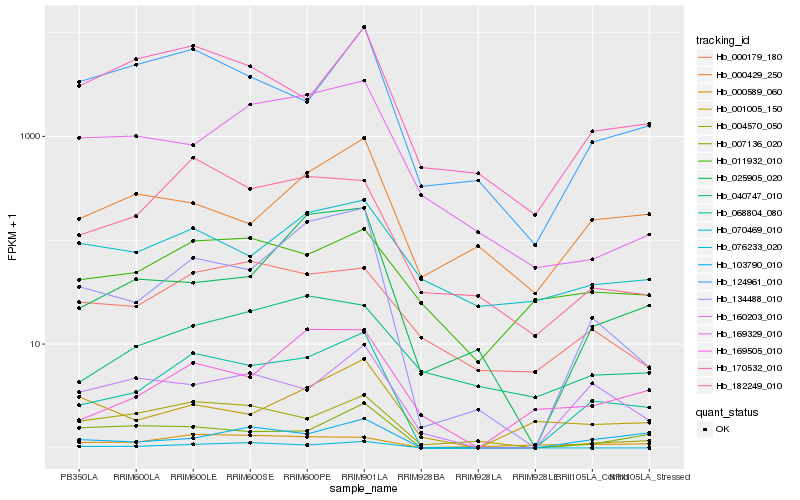
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068804_080 |
0.0 |
- |
- |
PREDICTED: protein BRICK 1 [Jatropha curcas] |
| 2 |
Hb_134488_010 |
0.1779364679 |
- |
- |
- |
| 3 |
Hb_160203_010 |
0.2153137185 |
- |
- |
- |
| 4 |
Hb_103790_010 |
0.2175671034 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40A isoform X4 [Jatropha curcas] |
| 5 |
Hb_007136_020 |
0.2176023642 |
- |
- |
PREDICTED: oxygen-dependent coproporphyrinogen-III oxidase, chloroplastic [Populus euphratica] |
| 6 |
Hb_004570_050 |
0.2195918474 |
- |
- |
kinase, putative [Ricinus communis] |
| 7 |
Hb_182249_010 |
0.2272010127 |
- |
- |
- |
| 8 |
Hb_124961_010 |
0.2288489763 |
- |
- |
hypothetical protein EUGRSUZ_B03893 [Eucalyptus grandis] |
| 9 |
Hb_170532_010 |
0.2301527354 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 8 [Jatropha curcas] |
| 10 |
Hb_169505_010 |
0.232583787 |
- |
- |
hypothetical protein JCGZ_04480 [Jatropha curcas] |
| 11 |
Hb_025905_020 |
0.2396982111 |
- |
- |
major allergen Pru ar 1-like [Jatropha curcas] |
| 12 |
Hb_000589_060 |
0.2422252896 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_00699 [Jatropha curcas] |
| 13 |
Hb_076233_020 |
0.2444335119 |
transcription factor |
TF Family: C2H2 |
PREDICTED: dnaJ homolog subfamily C member 21 [Populus euphratica] |
| 14 |
Hb_011932_010 |
0.2487002832 |
- |
- |
PREDICTED: probable calcium-binding protein CML49 [Jatropha curcas] |
| 15 |
Hb_000179_180 |
0.2490778963 |
- |
- |
hypothetical protein EUGRSUZ_C031461, partial [Eucalyptus grandis] |
| 16 |
Hb_040747_010 |
0.2553313976 |
- |
- |
hypothetical protein POPTR_0009s15120g [Populus trichocarpa] |
| 17 |
Hb_000429_250 |
0.2575615863 |
- |
- |
hypothetical protein L484_000744 [Morus notabilis] |
| 18 |
Hb_070469_010 |
0.2621251151 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 19 |
Hb_001005_150 |
0.2632947314 |
- |
- |
- |
| 20 |
Hb_169329_010 |
0.2653506407 |
- |
- |
PREDICTED: probable ADP-ribosylation factor GTPase-activating protein AGD14 isoform X2 [Jatropha curcas] |