| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
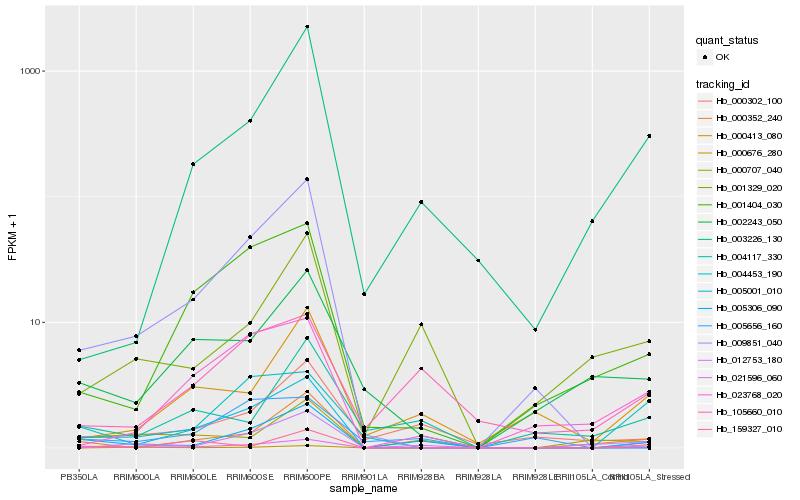
Hb_021596_060 |
0.0 |
- |
- |
PREDICTED: peroxidase 56-like, partial [Solanum tuberosum] |
| 2 |
Hb_005306_090 |
0.160033708 |
- |
- |
- |
| 3 |
Hb_000676_280 |
0.2232385343 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_012753_180 |
0.2400038365 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 5 |
Hb_000707_040 |
0.2567116878 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 6 |
Hb_001329_020 |
0.2625863381 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 7 |
Hb_004453_190 |
0.2660685971 |
- |
- |
- |
| 8 |
Hb_005001_010 |
0.2667254074 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 9 |
Hb_000352_240 |
0.2716513939 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 10 |
Hb_000413_080 |
0.274859248 |
- |
- |
- |
| 11 |
Hb_000302_100 |
0.2758109579 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 12 |
Hb_003226_130 |
0.279037127 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 13 |
Hb_023768_020 |
0.2810471303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001404_030 |
0.283146883 |
- |
- |
DWNN domain isoform 3 [Theobroma cacao] |
| 15 |
Hb_009851_040 |
0.2896510376 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_105660_010 |
0.290004881 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 17 |
Hb_002243_050 |
0.2947170831 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_004117_330 |
0.2956639556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005656_160 |
0.2957127193 |
- |
- |
PREDICTED: uncharacterized protein LOC105637448 [Jatropha curcas] |
| 20 |
Hb_159327_010 |
0.2962538576 |
- |
- |
hypothetical protein JCGZ_23768 [Jatropha curcas] |