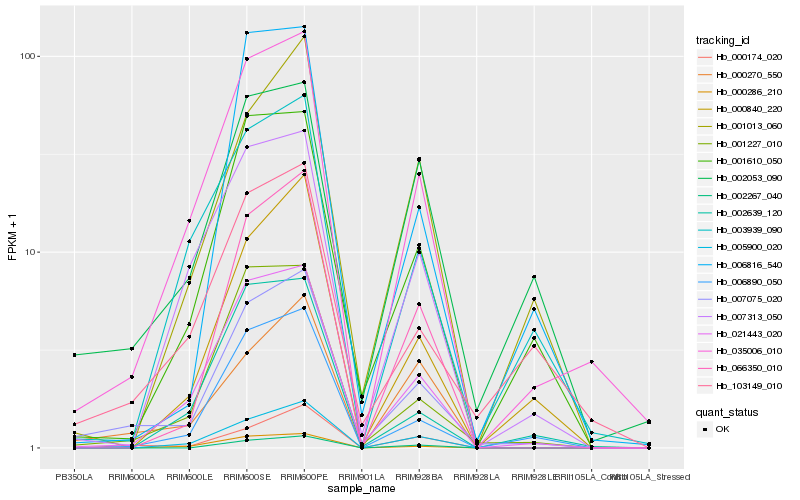
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007075_020 |
0.0 |
transcription factor |
TF Family: TRAF |
ankyrin repeat family protein [Populus trichocarpa] |
| 2 |
Hb_021443_020 |
0.1330275139 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 3 |
Hb_001227_010 |
0.1353076815 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0015s03140g [Populus trichocarpa] |
| 4 |
Hb_035006_010 |
0.1369307288 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 5 |
Hb_000174_020 |
0.1489487861 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 6 |
Hb_006890_050 |
0.1524813294 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor CYCLOIDEA [Jatropha curcas] |
| 7 |
Hb_103149_010 |
0.1570130438 |
- |
- |
hypothetical protein CISIN_1g0397312mg, partial [Citrus sinensis] |
| 8 |
Hb_005900_020 |
0.1589667058 |
- |
- |
- |
| 9 |
Hb_066350_010 |
0.1599452858 |
- |
- |
somatic embryogenesis receptor-like kinase 4, partial [Rosa hybrid cultivar] |
| 10 |
Hb_001610_050 |
0.1601591029 |
- |
- |
PREDICTED: subtilisin-like protease-like [Solanum tuberosum] |
| 11 |
Hb_002053_090 |
0.1665498238 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000840_220 |
0.1668363419 |
- |
- |
PREDICTED: RING-H2 finger protein ATL78-like [Jatropha curcas] |
| 13 |
Hb_000270_550 |
0.17046012 |
- |
- |
- |
| 14 |
Hb_007313_050 |
0.1769989648 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 15 |
Hb_000286_210 |
0.1791962005 |
- |
- |
hypothetical protein EUGRSUZ_C00018 [Eucalyptus grandis] |
| 16 |
Hb_001013_060 |
0.1804938366 |
- |
- |
- |
| 17 |
Hb_006816_540 |
0.1858223102 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET16-like [Jatropha curcas] |
| 18 |
Hb_002639_120 |
0.1863970922 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_003939_090 |
0.1871611371 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 20 |
Hb_002267_040 |
0.1876598985 |
- |
- |
- |