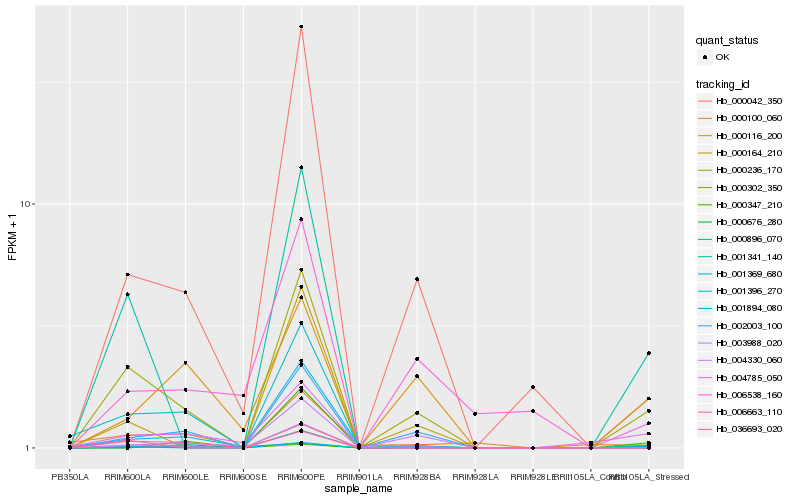
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_170 |
0.0 |
- |
- |
PREDICTED: Werner Syndrome-like exonuclease [Jatropha curcas] |
| 2 |
Hb_000042_350 |
0.2221651352 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 3 |
Hb_003988_020 |
0.2384110724 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 4 |
Hb_001894_080 |
0.2408519927 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_002003_100 |
0.24435831 |
- |
- |
- |
| 6 |
Hb_001341_140 |
0.2535443146 |
- |
- |
- |
| 7 |
Hb_006663_110 |
0.2917157429 |
- |
- |
PREDICTED: U-box domain-containing protein 34-like [Camelina sativa] |
| 8 |
Hb_000302_350 |
0.2943201446 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 9 |
Hb_000676_280 |
0.2947290897 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_001396_270 |
0.2978515884 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 11 |
Hb_000116_200 |
0.2978828444 |
transcription factor |
TF Family: HMG |
hypothetical protein POPTR_0005s10440g [Populus trichocarpa] |
| 12 |
Hb_004785_050 |
0.2992428982 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 13 |
Hb_001369_680 |
0.3010141994 |
- |
- |
PREDICTED: uncharacterized protein LOC105648582 [Jatropha curcas] |
| 14 |
Hb_000164_210 |
0.3017358816 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 15 |
Hb_006538_160 |
0.3046616671 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000100_060 |
0.3089084024 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 17 |
Hb_004330_060 |
0.3092658029 |
- |
- |
hypothetical protein POPTR_0011s01530g [Populus trichocarpa] |
| 18 |
Hb_000896_070 |
0.3107442178 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 19 |
Hb_036693_020 |
0.3150608441 |
- |
- |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 20 |
Hb_000347_210 |
0.3178541229 |
- |
- |
PREDICTED: uncharacterized protein LOC105631371 [Jatropha curcas] |