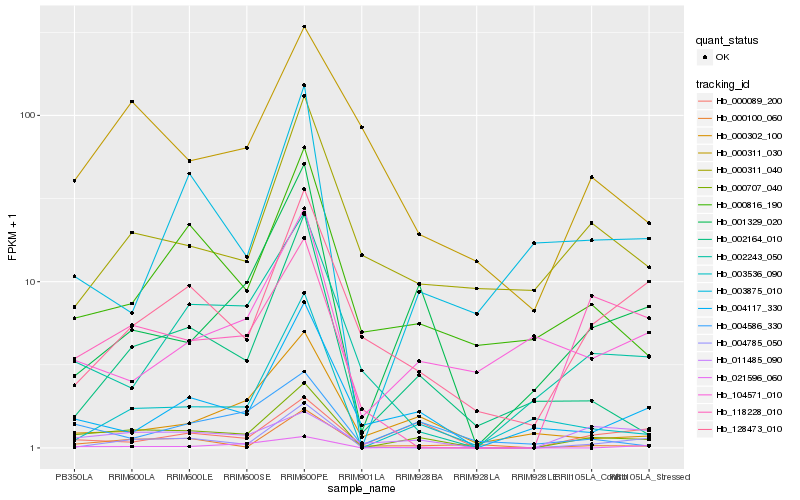
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000707_040 |
0.0 |
- |
- |
Uncharacterized protein TCM_021829 [Theobroma cacao] |
| 2 |
Hb_001329_020 |
0.2130475967 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 3 |
Hb_002164_010 |
0.2399089039 |
- |
- |
Stellacyanin, putative [Ricinus communis] |
| 4 |
Hb_000816_190 |
0.2507016205 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_003536_090 |
0.2509953247 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_004117_330 |
0.2520427995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000089_200 |
0.2548840745 |
- |
- |
hypothetical protein VITISV_019292 [Vitis vinifera] |
| 8 |
Hb_000100_060 |
0.2564550762 |
- |
- |
PREDICTED: probable glucan endo-1,3-beta-glucosidase A6 [Populus euphratica] |
| 9 |
Hb_021596_060 |
0.2567116878 |
- |
- |
PREDICTED: peroxidase 56-like, partial [Solanum tuberosum] |
| 10 |
Hb_000302_100 |
0.2604513582 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 11 |
Hb_011485_090 |
0.2619576705 |
- |
- |
- |
| 12 |
Hb_004586_330 |
0.2621032917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_128473_010 |
0.2625823476 |
- |
- |
- |
| 14 |
Hb_004785_050 |
0.266005435 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 15 |
Hb_002243_050 |
0.2674910043 |
- |
- |
PREDICTED: ion channel DMI1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_118228_010 |
0.2731499453 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 17 |
Hb_003875_010 |
0.2750973584 |
- |
- |
hypothetical protein POPTR_0002s14490g, partial [Populus trichocarpa] |
| 18 |
Hb_000311_040 |
0.2766077915 |
- |
- |
PREDICTED: S-formylglutathione hydrolase [Jatropha curcas] |
| 19 |
Hb_000311_030 |
0.2818647134 |
- |
- |
hypothetical protein B456_002G227600 [Gossypium raimondii] |
| 20 |
Hb_104571_010 |
0.2847435249 |
- |
- |
PREDICTED: metal transporter Nramp1 [Jatropha curcas] |