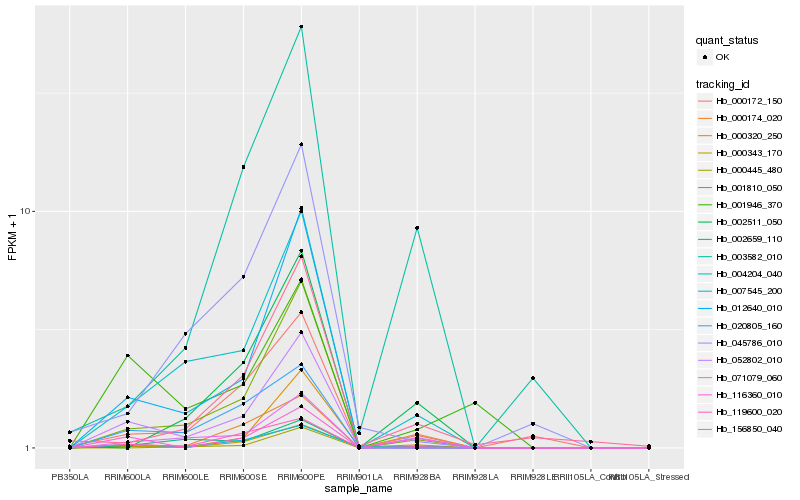
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_052802_010 |
0.0 |
- |
- |
kinase, putative [Ricinus communis] |
| 2 |
Hb_001810_050 |
0.1203225726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004204_040 |
0.1464004911 |
- |
- |
- |
| 4 |
Hb_007545_200 |
0.1603031424 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 5 |
Hb_012640_010 |
0.1679897216 |
- |
- |
- |
| 6 |
Hb_119600_020 |
0.174721096 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_071079_060 |
0.1837165209 |
- |
- |
PREDICTED: uncharacterized protein LOC105113464 [Populus euphratica] |
| 8 |
Hb_000172_150 |
0.1925074345 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Populus euphratica] |
| 9 |
Hb_116360_010 |
0.2017967618 |
- |
- |
- |
| 10 |
Hb_000343_170 |
0.2043280612 |
- |
- |
PREDICTED: probable sulfate transporter 3.4 [Jatropha curcas] |
| 11 |
Hb_020805_160 |
0.2069128856 |
- |
- |
- |
| 12 |
Hb_002659_110 |
0.2081265707 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_000320_250 |
0.2175986418 |
- |
- |
hypothetical protein POPTR_0021s00440g [Populus trichocarpa] |
| 14 |
Hb_000174_020 |
0.2301619714 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 15 |
Hb_000445_480 |
0.2351277657 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_002511_050 |
0.2368618254 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_156850_040 |
0.2380056437 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 18 |
Hb_003582_010 |
0.2381131108 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 19 |
Hb_001946_370 |
0.2387065105 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_045786_010 |
0.2402032301 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |