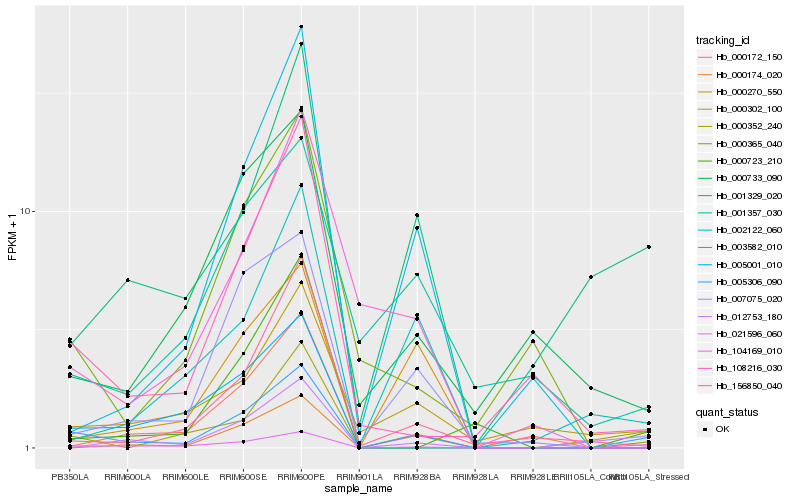
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005306_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_021596_060 |
0.160033708 |
- |
- |
PREDICTED: peroxidase 56-like, partial [Solanum tuberosum] |
| 3 |
Hb_000302_100 |
0.2499820044 |
- |
- |
PREDICTED: uncharacterized protein LOC105766209 [Gossypium raimondii] |
| 4 |
Hb_001329_020 |
0.2533517877 |
- |
- |
PREDICTED: probable boron transporter 2 [Populus euphratica] |
| 5 |
Hb_108216_030 |
0.2590573015 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_012753_180 |
0.2595552619 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 7 |
Hb_007075_020 |
0.2596243118 |
transcription factor |
TF Family: TRAF |
ankyrin repeat family protein [Populus trichocarpa] |
| 8 |
Hb_002122_060 |
0.2693972624 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_000270_550 |
0.2705285005 |
- |
- |
- |
| 10 |
Hb_000174_020 |
0.2721313661 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 11 |
Hb_156850_040 |
0.2741137445 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 12 |
Hb_003582_010 |
0.278592985 |
- |
- |
hypothetical protein EUGRSUZ_B026451, partial [Eucalyptus grandis] |
| 13 |
Hb_000172_150 |
0.280034997 |
- |
- |
PREDICTED: protein UPSTREAM OF FLC [Populus euphratica] |
| 14 |
Hb_000733_090 |
0.2801883246 |
- |
- |
PREDICTED: putative lipid-transfer protein DIR1 [Jatropha curcas] |
| 15 |
Hb_000723_210 |
0.2808947326 |
- |
- |
Indole-3-acetic acid-induced protein ARG7, putative [Ricinus communis] |
| 16 |
Hb_001357_030 |
0.2815807977 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 17 |
Hb_000352_240 |
0.2822017009 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 18 |
Hb_005001_010 |
0.2823992038 |
- |
- |
GAST1 protein precursor, putative [Ricinus communis] |
| 19 |
Hb_104169_010 |
0.2835057806 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 20 |
Hb_000365_040 |
0.2835169294 |
- |
- |
PREDICTED: uncharacterized protein LOC105649097 [Jatropha curcas] |