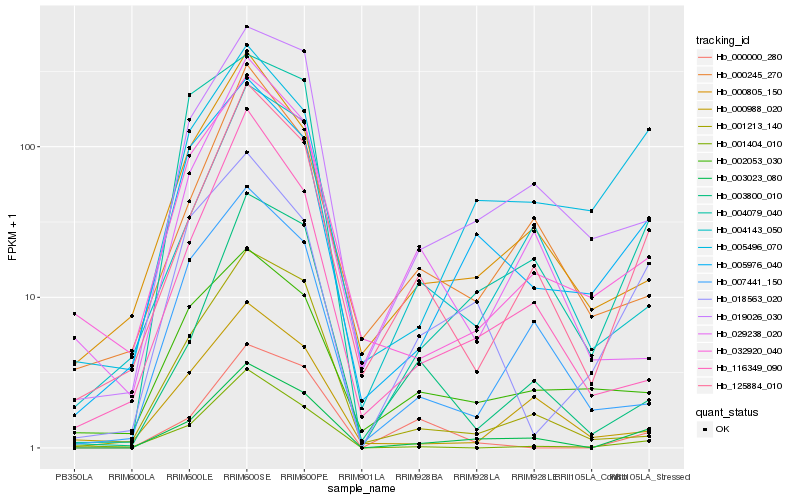
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003023_080 |
0.0 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_125884_010 |
0.1395632561 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Jatropha curcas] |
| 3 |
Hb_019026_030 |
0.148721764 |
- |
- |
hypothetical protein JCGZ_00744 [Jatropha curcas] |
| 4 |
Hb_005976_040 |
0.1581825275 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like [Jatropha curcas] |
| 5 |
Hb_004143_050 |
0.1743979398 |
- |
- |
PREDICTED: uncharacterized protein LOC105630712 [Jatropha curcas] |
| 6 |
Hb_032920_040 |
0.1844746178 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s30260g [Populus trichocarpa] |
| 7 |
Hb_000805_150 |
0.1852685818 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_004079_040 |
0.1861757101 |
- |
- |
hypothetical protein JCGZ_07227 [Jatropha curcas] |
| 9 |
Hb_001213_140 |
0.1906284862 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 10 |
Hb_018563_020 |
0.1939411778 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 6 [Hevea brasiliensis] |
| 11 |
Hb_007441_150 |
0.1939948477 |
- |
- |
Ankyrin repeat family protein [Theobroma cacao] |
| 12 |
Hb_001404_010 |
0.1950534755 |
- |
- |
hypothetical protein JCGZ_22818 [Jatropha curcas] |
| 13 |
Hb_000245_270 |
0.1954769613 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 14 |
Hb_116349_090 |
0.1958482575 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 9 [Jatropha curcas] |
| 15 |
Hb_005496_070 |
0.1985454149 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT10-like [Jatropha curcas] |
| 16 |
Hb_003800_010 |
0.1990742551 |
- |
- |
PREDICTED: cysteine-rich and transmembrane domain-containing protein A-like [Jatropha curcas] |
| 17 |
Hb_000988_020 |
0.2034772884 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000000_280 |
0.2036642895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_029238_020 |
0.2057648104 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 20 |
Hb_002053_030 |
0.2072452727 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |