| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
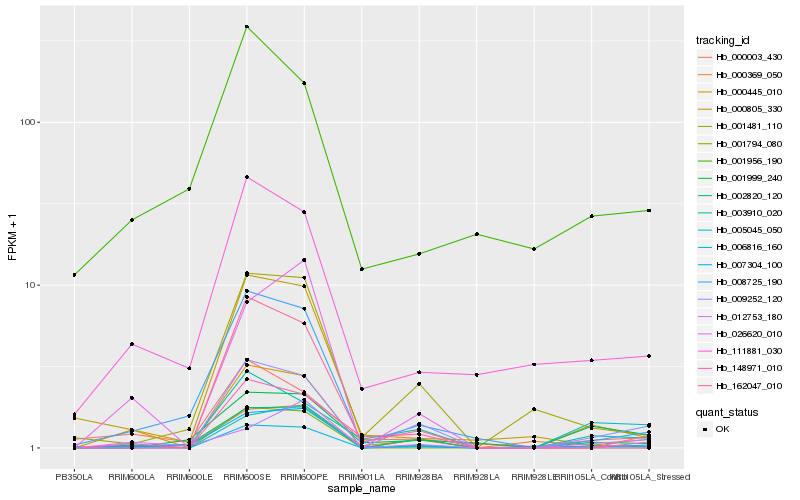
Hb_000445_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |
| 2 |
Hb_006816_160 |
0.1926169751 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 3 |
Hb_009252_120 |
0.2315940377 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002820_120 |
0.2402399178 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 5 |
Hb_111881_030 |
0.241611953 |
- |
- |
PREDICTED: uncharacterized protein LOC105645255 [Jatropha curcas] |
| 6 |
Hb_003910_020 |
0.2424581036 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 7 |
Hb_008725_190 |
0.2518108281 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 5-like [Jatropha curcas] |
| 8 |
Hb_000003_430 |
0.2525638303 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 9 |
Hb_007304_100 |
0.2700849084 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Jatropha curcas] |
| 10 |
Hb_001794_080 |
0.2783707131 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_148971_010 |
0.2824381082 |
- |
- |
Disease resistance protein RPS5, putative [Ricinus communis] |
| 12 |
Hb_162047_010 |
0.2824396841 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 13 |
Hb_001999_240 |
0.283239146 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 14 |
Hb_001481_110 |
0.2842393056 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 15 |
Hb_012753_180 |
0.2871560951 |
- |
- |
PREDICTED: cytochrome P450 87A3 [Jatropha curcas] |
| 16 |
Hb_000805_330 |
0.2885924015 |
- |
- |
PREDICTED: ricin-like [Elaeis guineensis] |
| 17 |
Hb_001956_190 |
0.2913677276 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 18 |
Hb_005045_050 |
0.2931672359 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 19 |
Hb_026620_010 |
0.2945844409 |
- |
- |
- |
| 20 |
Hb_000369_050 |
0.2947353516 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |