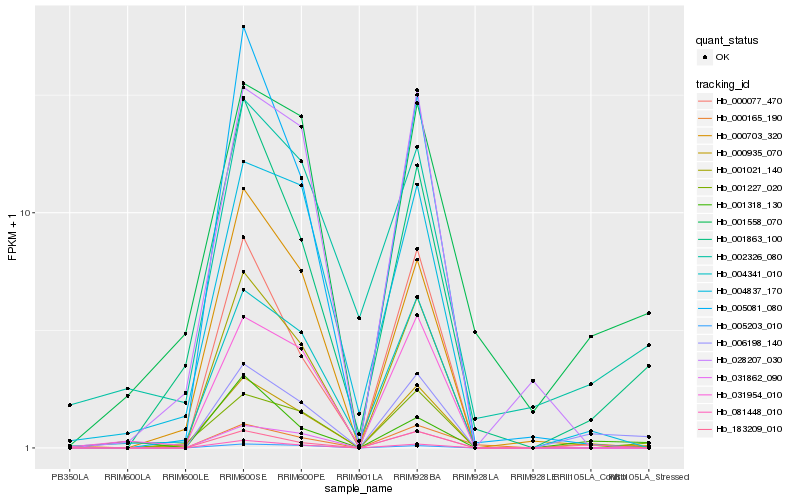
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006198_140 |
0.0 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 2 |
Hb_183209_010 |
0.1610305477 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000935_070 |
0.1725657528 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 4 |
Hb_001227_020 |
0.1967173912 |
- |
- |
- |
| 5 |
Hb_001558_070 |
0.1970049666 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 6 |
Hb_001021_140 |
0.2019396472 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 7 |
Hb_001863_100 |
0.2070954989 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_031862_090 |
0.220631154 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 9 |
Hb_001318_130 |
0.2211807998 |
- |
- |
PREDICTED: short-chain dehydrogenase reductase 3b-like [Populus euphratica] |
| 10 |
Hb_000165_190 |
0.2214739109 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 11 |
Hb_002326_080 |
0.2271921814 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.6 [Jatropha curcas] |
| 12 |
Hb_081448_010 |
0.2274936331 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 13 |
Hb_031954_010 |
0.2323824219 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_004341_010 |
0.2330622767 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 15 |
Hb_000703_320 |
0.2347391669 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 16 |
Hb_000077_470 |
0.2357459732 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 17 |
Hb_028207_030 |
0.2374205618 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 18 |
Hb_005203_010 |
0.2382484334 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 19 |
Hb_004837_170 |
0.2387877778 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 20 |
Hb_005081_080 |
0.2399559589 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |