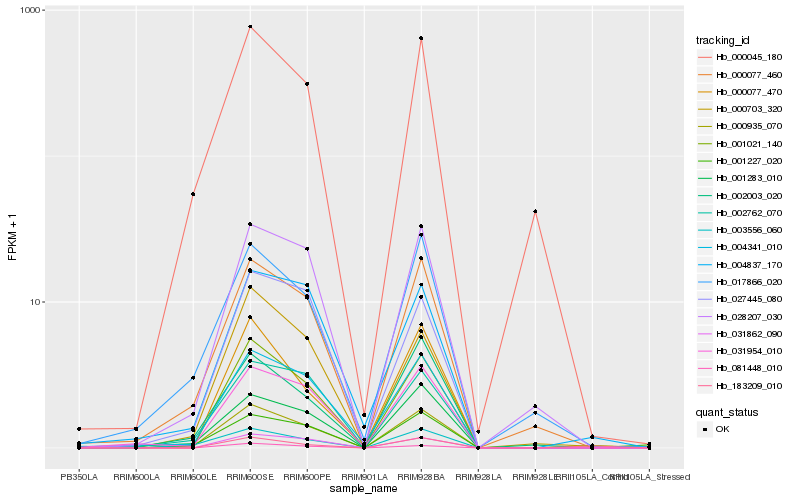
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000935_070 |
0.0 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001021_140 |
0.1163110939 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 3 |
Hb_000077_460 |
0.1310766414 |
- |
- |
- |
| 4 |
Hb_004341_010 |
0.137906114 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 5 |
Hb_003556_060 |
0.1409031222 |
- |
- |
- |
| 6 |
Hb_001283_010 |
0.1409224402 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 7 |
Hb_028207_030 |
0.1424532553 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 8 |
Hb_031954_010 |
0.1452151411 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_002762_070 |
0.1484977575 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 10 |
Hb_031862_090 |
0.1497108751 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 11 |
Hb_000703_320 |
0.1505073828 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 12 |
Hb_183209_010 |
0.1514290914 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000045_180 |
0.1519208318 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 14 |
Hb_004837_170 |
0.1570056807 |
- |
- |
PREDICTED: uncharacterized protein LOC105648284 [Jatropha curcas] |
| 15 |
Hb_081448_010 |
0.1573945124 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 16 |
Hb_017866_020 |
0.1591659795 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 40 [Jatropha curcas] |
| 17 |
Hb_000077_470 |
0.1613557195 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 18 |
Hb_002003_020 |
0.161990733 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 19 |
Hb_027445_080 |
0.1628268441 |
transcription factor |
TF Family: LOB |
LOB domain protein 25 [Populus trichocarpa] |
| 20 |
Hb_001227_020 |
0.1631234963 |
- |
- |
- |