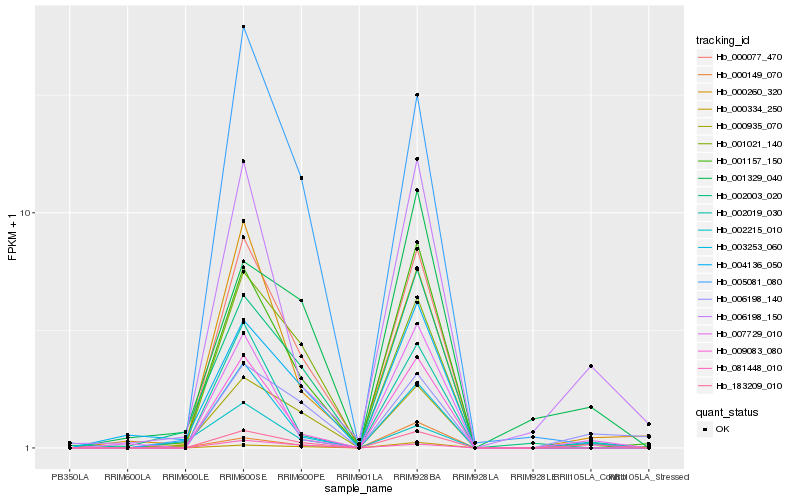
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183209_010 |
0.0 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g63930 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000935_070 |
0.1514290914 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 3 |
Hb_006198_140 |
0.1610305477 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 4 |
Hb_000077_470 |
0.1950956059 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 5 |
Hb_006198_150 |
0.2040448595 |
- |
- |
- |
| 6 |
Hb_001021_140 |
0.2090887482 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 7 |
Hb_002019_030 |
0.2186588482 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 8 |
Hb_001157_150 |
0.2203759144 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 9 |
Hb_001329_040 |
0.2213344482 |
- |
- |
hypothetical protein RCOM_1345340 [Ricinus communis] |
| 10 |
Hb_005081_080 |
0.2234311431 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 11 |
Hb_009083_080 |
0.2242227674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000260_320 |
0.2264579629 |
- |
- |
PREDICTED: uncharacterized protein LOC105649058 [Jatropha curcas] |
| 13 |
Hb_007729_010 |
0.2287649078 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 14 |
Hb_003253_060 |
0.2317852728 |
- |
- |
PREDICTED: tropinone reductase-like 1 [Vitis vinifera] |
| 15 |
Hb_002003_020 |
0.2318251086 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 16 |
Hb_000149_070 |
0.2333065981 |
- |
- |
PREDICTED: uncharacterized protein At5g39570 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004136_050 |
0.2334975305 |
- |
- |
- |
| 18 |
Hb_000334_250 |
0.2349639277 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_081448_010 |
0.2373257012 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 20 |
Hb_002215_010 |
0.238005216 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |