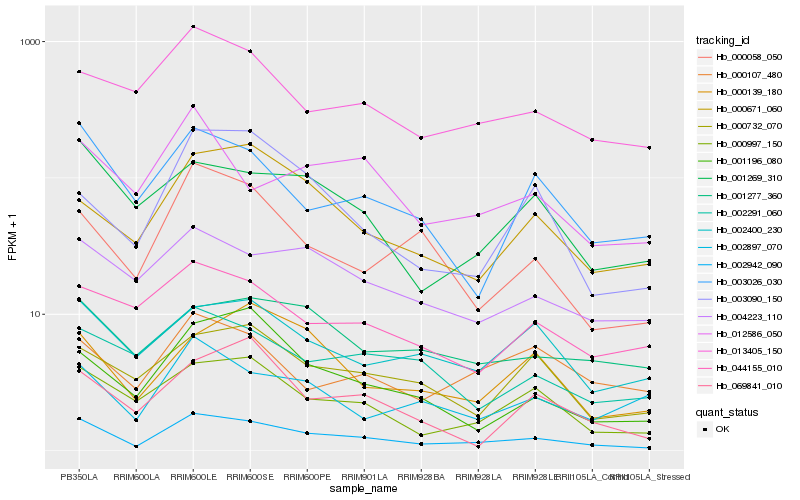
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_090 |
0.0 |
- |
- |
DNAJ heat shock N-terminal domain-containing protein, putative [Theobroma cacao] |
| 2 |
Hb_000997_150 |
0.1513801552 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_000058_050 |
0.1671091274 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 4 |
Hb_003026_030 |
0.1672840645 |
- |
- |
transformer serine/arginine-rich ribonucleoprotein [Populus trichocarpa] |
| 5 |
Hb_002400_230 |
0.170317391 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 6 |
Hb_001269_310 |
0.1712889911 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 6 [Jatropha curcas] |
| 7 |
Hb_004223_110 |
0.1726669161 |
- |
- |
PREDICTED: putative HVA22-like protein g [Populus euphratica] |
| 8 |
Hb_013405_150 |
0.1735867128 |
- |
- |
PREDICTED: actin cytoskeleton-regulatory complex protein pan1 [Jatropha curcas] |
| 9 |
Hb_001196_080 |
0.1738812755 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 10 |
Hb_000671_060 |
0.1739331289 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_002897_070 |
0.1799904563 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 12 |
Hb_000732_070 |
0.1801570538 |
- |
- |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 13 |
Hb_001277_360 |
0.181645885 |
- |
- |
PREDICTED: dymeclin isoform X1 [Jatropha curcas] |
| 14 |
Hb_000139_180 |
0.181947291 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14440 [Jatropha curcas] |
| 15 |
Hb_000107_480 |
0.1821368541 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 6-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_003090_150 |
0.1822069905 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 17 |
Hb_002291_060 |
0.1846361591 |
- |
- |
PREDICTED: splicing factor 3A subunit 3-like isoform X1 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_044155_010 |
0.1864003873 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_069841_010 |
0.1922912022 |
- |
- |
resistance protein [Quercus suber] |
| 20 |
Hb_012586_050 |
0.1924671975 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Prunus mume] |