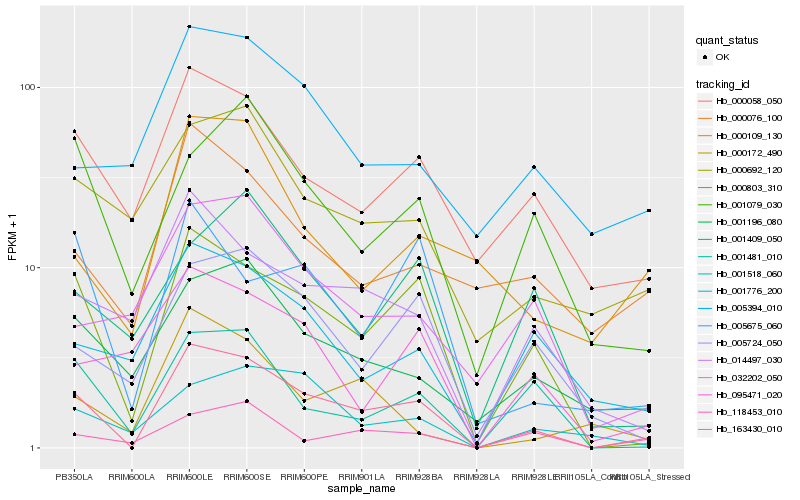
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163430_010 |
0.0 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 2 |
Hb_000803_310 |
0.1552214916 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105648332 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001481_010 |
0.2221556178 |
- |
- |
PREDICTED: uncharacterized protein LOC105632300 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000058_050 |
0.2241525863 |
- |
- |
PREDICTED: uncharacterized protein LOC105640877 [Jatropha curcas] |
| 5 |
Hb_005724_050 |
0.2291496403 |
- |
- |
- |
| 6 |
Hb_001196_080 |
0.2323317778 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 7 |
Hb_014497_030 |
0.2323862839 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 8 |
Hb_000692_120 |
0.2402358129 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 9 |
Hb_001776_200 |
0.2411505909 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 2 isoform X2 [Pyrus x bretschneideri] |
| 10 |
Hb_005675_060 |
0.2446312814 |
- |
- |
PREDICTED: protein NIM1-INTERACTING 2 [Jatropha curcas] |
| 11 |
Hb_001079_030 |
0.2471878611 |
- |
- |
PREDICTED: splicing factor U2af small subunit B-like [Jatropha curcas] |
| 12 |
Hb_000076_100 |
0.2495030957 |
- |
- |
PREDICTED: septum-promoting GTP-binding protein 1 [Jatropha curcas] |
| 13 |
Hb_118453_010 |
0.2496209524 |
- |
- |
hypothetical protein VITISV_012360 [Vitis vinifera] |
| 14 |
Hb_001518_060 |
0.2515544172 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 9 [Jatropha curcas] |
| 15 |
Hb_095471_020 |
0.2520493959 |
- |
- |
- |
| 16 |
Hb_032202_050 |
0.2534819064 |
transcription factor |
TF Family: TCP |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_005394_010 |
0.2543113659 |
- |
- |
PREDICTED: calmodulin-binding protein 25-like [Jatropha curcas] |
| 18 |
Hb_000172_490 |
0.2553697043 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 19 |
Hb_000109_130 |
0.2554751572 |
- |
- |
PREDICTED: probable alpha,alpha-trehalose-phosphate synthase [UDP-forming] 9 [Jatropha curcas] |
| 20 |
Hb_001409_050 |
0.2603653346 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |