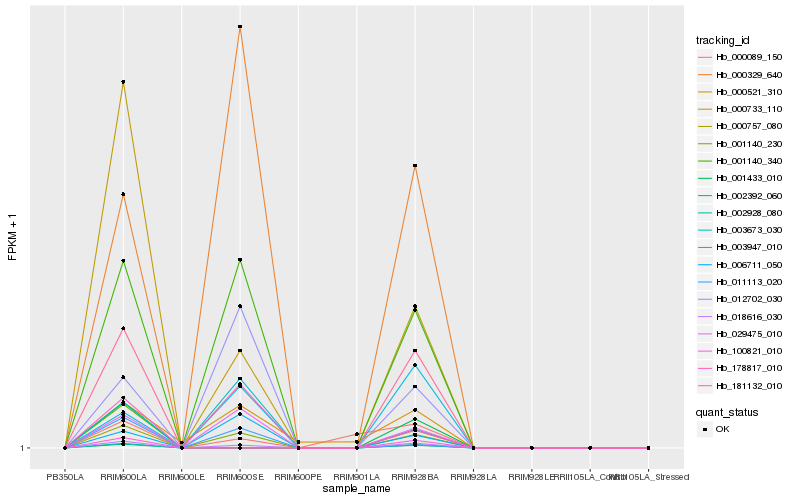
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018616_030 |
0.0 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 2 |
Hb_001140_230 |
0.1529379091 |
- |
- |
- |
| 3 |
Hb_001140_340 |
0.170776856 |
- |
- |
- |
| 4 |
Hb_000733_110 |
0.1911036612 |
- |
- |
hypothetical protein JCGZ_01360 [Jatropha curcas] |
| 5 |
Hb_029475_010 |
0.2222769178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003947_010 |
0.259359067 |
- |
- |
hypothetical protein JCGZ_24829 [Jatropha curcas] |
| 7 |
Hb_000521_310 |
0.2808144401 |
- |
- |
- |
| 8 |
Hb_000329_640 |
0.2897530916 |
- |
- |
- |
| 9 |
Hb_178817_010 |
0.2925623207 |
- |
- |
protease [Gossypium hirsutum] |
| 10 |
Hb_006711_050 |
0.3090512424 |
- |
- |
PREDICTED: putative germin-like protein 2-1 [Jatropha curcas] |
| 11 |
Hb_012702_030 |
0.3144422443 |
- |
- |
hypothetical protein POPTR_0077s002001g, partial [Populus trichocarpa] |
| 12 |
Hb_003673_030 |
0.3156629932 |
- |
- |
hypothetical protein POPTR_0011s11940g [Populus trichocarpa] |
| 13 |
Hb_011113_020 |
0.3168502237 |
- |
- |
hypothetical protein CICLE_v10018209mg [Citrus clementina] |
| 14 |
Hb_000089_150 |
0.3195791225 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 15 |
Hb_002392_060 |
0.3235625028 |
- |
- |
gag/pol protein [Bryonia dioica] |
| 16 |
Hb_002928_080 |
0.32356373 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g42310, mitochondrial [Jatropha curcas] |
| 17 |
Hb_100821_010 |
0.323616638 |
- |
- |
hypothetical protein JCGZ_01613 [Jatropha curcas] |
| 18 |
Hb_000757_080 |
0.3240127448 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |
| 19 |
Hb_181132_010 |
0.3242028268 |
- |
- |
- |
| 20 |
Hb_001433_010 |
0.3246271362 |
- |
- |
- |