| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
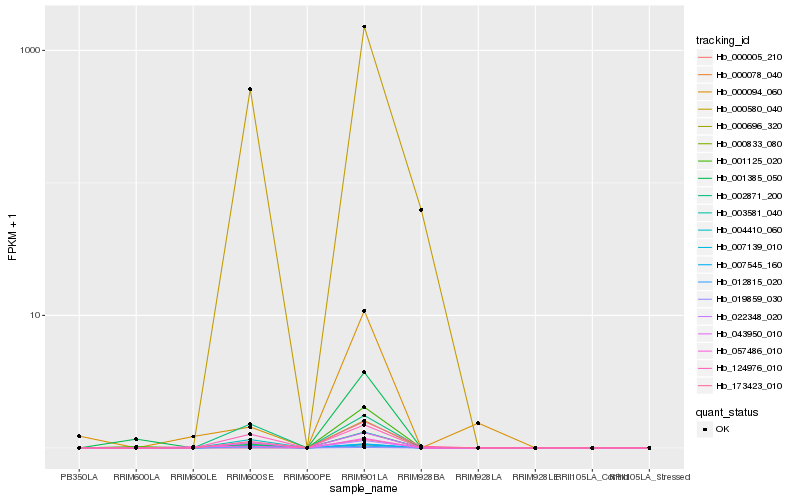
Hb_000833_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105789525 [Gossypium raimondii] |
| 2 |
Hb_000696_320 |
0.0289926829 |
- |
- |
PREDICTED: uncharacterized protein LOC104884717 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_043950_010 |
0.1129193745 |
- |
- |
PREDICTED: uncharacterized protein LOC102619199, partial [Citrus sinensis] |
| 4 |
Hb_173423_010 |
0.1489537014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007545_160 |
0.1563918894 |
transcription factor |
TF Family: HB |
Protein WUSCHEL, putative [Ricinus communis] |
| 6 |
Hb_000580_040 |
0.1699021938 |
- |
- |
- |
| 7 |
Hb_001125_020 |
0.1790578447 |
- |
- |
hypothetical protein B456_001G219500 [Gossypium raimondii] |
| 8 |
Hb_004410_060 |
0.1915823404 |
- |
- |
- |
| 9 |
Hb_124976_010 |
0.2113603259 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 10 |
Hb_019859_030 |
0.2182912662 |
- |
- |
hypothetical protein MIMGU_mgv1a018902mg [Erythranthe guttata] |
| 11 |
Hb_007139_010 |
0.21857282 |
- |
- |
hypothetical protein POPTR_0016s134801g, partial [Populus trichocarpa] |
| 12 |
Hb_012815_020 |
0.2192859519 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_057486_010 |
0.2229744301 |
- |
- |
PREDICTED: uncharacterized protein LOC105793230 [Gossypium raimondii] |
| 14 |
Hb_000078_040 |
0.2329375963 |
- |
- |
PREDICTED: uncharacterized protein LOC104907366 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_022348_020 |
0.2517783955 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 16 |
Hb_003581_040 |
0.2538243287 |
- |
- |
hypothetical protein PHAVU_002G080900g [Phaseolus vulgaris] |
| 17 |
Hb_001385_050 |
0.2543053746 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_002871_200 |
0.2548607277 |
- |
- |
- |
| 19 |
Hb_000094_060 |
0.2571105189 |
- |
- |
- |
| 20 |
Hb_000005_210 |
0.2729480716 |
- |
- |
- |