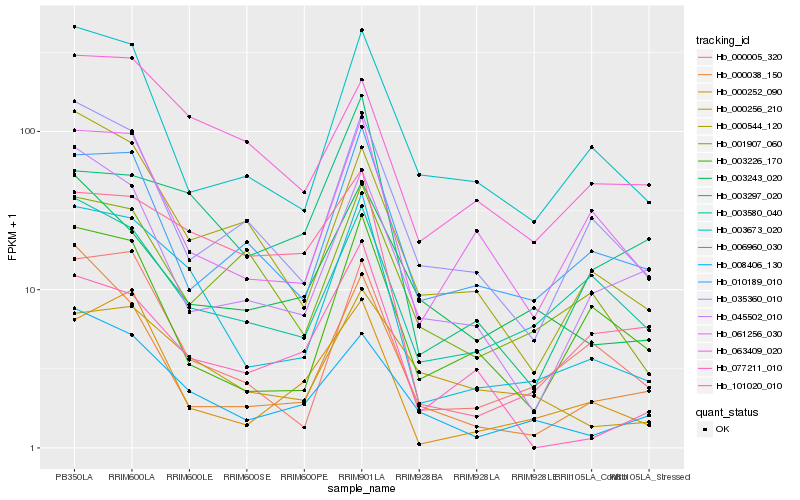
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006960_030 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB44-like [Jatropha curcas] |
| 2 |
Hb_000005_320 |
0.1704985823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_008406_130 |
0.170879221 |
- |
- |
- |
| 4 |
Hb_061256_030 |
0.1860123139 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 5 |
Hb_003673_020 |
0.1867170136 |
- |
- |
hypothetical protein JCGZ_20998 [Jatropha curcas] |
| 6 |
Hb_101020_010 |
0.1881849516 |
- |
- |
- |
| 7 |
Hb_000252_090 |
0.1910894687 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 8 |
Hb_000544_120 |
0.1926221262 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 9 |
Hb_035360_010 |
0.195189905 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 10 |
Hb_000256_210 |
0.1959201708 |
- |
- |
PREDICTED: 40S ribosomal protein S11-like [Phoenix dactylifera] |
| 11 |
Hb_063409_020 |
0.1969466157 |
- |
- |
Calcium-binding protein, putative [Ricinus communis] |
| 12 |
Hb_003226_170 |
0.1975093292 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003580_040 |
0.2004045636 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2-like [Jatropha curcas] |
| 14 |
Hb_077211_010 |
0.201618662 |
- |
- |
PREDICTED: lariat debranching enzyme-like [Jatropha curcas] |
| 15 |
Hb_010189_010 |
0.2022773523 |
- |
- |
- |
| 16 |
Hb_003243_020 |
0.2093397357 |
- |
- |
PREDICTED: uncharacterized protein LOC105629849 [Jatropha curcas] |
| 17 |
Hb_045502_010 |
0.2106598632 |
- |
- |
prefoldin subunit 1 [Jatropha curcas] |
| 18 |
Hb_000038_150 |
0.2144633771 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 19 |
Hb_003297_020 |
0.2148257461 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 20 |
Hb_001907_060 |
0.2166152827 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 5 [Jatropha curcas] |