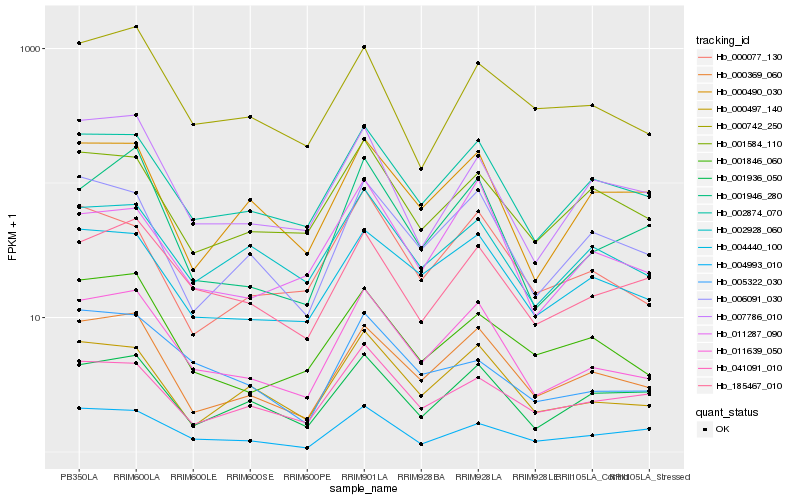
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011639_050 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000369_060 |
0.0871037277 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 3 |
Hb_002874_070 |
0.0947272127 |
- |
- |
PREDICTED: RNA-binding KH domain-containing protein PEPPER-like [Jatropha curcas] |
| 4 |
Hb_000497_140 |
0.103866355 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like [Jatropha curcas] |
| 5 |
Hb_004440_100 |
0.1086785681 |
- |
- |
PREDICTED: transmembrane protein 53 [Jatropha curcas] |
| 6 |
Hb_001946_280 |
0.1109896028 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_185467_010 |
0.1162214837 |
- |
- |
- |
| 8 |
Hb_006091_030 |
0.118460415 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog C-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001846_060 |
0.1248780073 |
- |
- |
hypothetical protein RCOM_0992860 [Ricinus communis] |
| 10 |
Hb_007786_010 |
0.1252863317 |
- |
- |
PREDICTED: uncharacterized protein LOC100801920 [Glycine max] |
| 11 |
Hb_001936_050 |
0.1268328757 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210-like [Prunus mume] |
| 12 |
Hb_000077_130 |
0.1276416086 |
- |
- |
PREDICTED: callose synthase 12 [Jatropha curcas] |
| 13 |
Hb_011287_090 |
0.128253347 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 14 |
Hb_002928_060 |
0.1285870752 |
- |
- |
PREDICTED: pre-rRNA-processing protein TSR1 homolog [Jatropha curcas] |
| 15 |
Hb_000742_250 |
0.1317411547 |
- |
- |
Tubulin beta-6 chain [Gossypium arboreum] |
| 16 |
Hb_000490_030 |
0.1320387658 |
- |
- |
DNA-directed RNA polymerase II subunit, putative [Ricinus communis] |
| 17 |
Hb_001584_110 |
0.1323774472 |
- |
- |
PREDICTED: importin subunit alpha-4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_041091_010 |
0.1345326398 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g05670, mitochondrial [Jatropha curcas] |
| 19 |
Hb_004993_010 |
0.1352638661 |
- |
- |
PREDICTED: uncharacterized protein LOC104889591 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_005322_030 |
0.1356594058 |
- |
- |
PREDICTED: rRNA-processing protein UTP23 homolog [Jatropha curcas] |