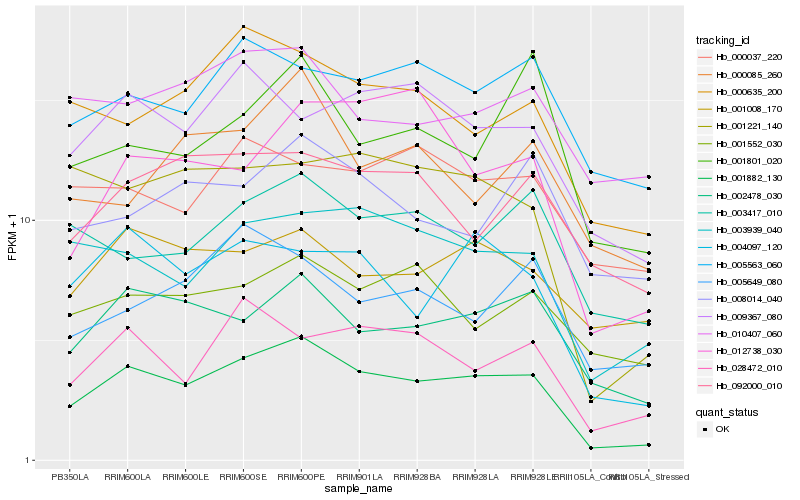
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001882_130 |
0.0 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028996 [Vitis vinifera] |
| 2 |
Hb_002478_030 |
0.123929842 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 3 |
Hb_004097_120 |
0.1263825065 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_003939_040 |
0.1301045154 |
- |
- |
hypothetical protein CICLE_v10014350mg [Citrus clementina] |
| 5 |
Hb_028472_010 |
0.138477249 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000635_200 |
0.1395917171 |
rubber biosynthesis |
Gene Name: SRPP5 |
PREDICTED: REF/SRPP-like protein At1g67360 [Jatropha curcas] |
| 7 |
Hb_001221_140 |
0.1416836628 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_009367_080 |
0.1432187065 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor 42 [Jatropha curcas] |
| 9 |
Hb_005563_060 |
0.1483962089 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 10 |
Hb_003417_010 |
0.148545877 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 11 |
Hb_092000_010 |
0.1497074147 |
- |
- |
kelch repeat-containing F-box family protein [Populus trichocarpa] |
| 12 |
Hb_001008_170 |
0.1527111938 |
transcription factor |
TF Family: mTERF |
RNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_001552_030 |
0.1537841475 |
- |
- |
PREDICTED: galactoside 2-alpha-L-fucosyltransferase-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_010407_060 |
0.1556916223 |
- |
- |
PREDICTED: probable methyltransferase PMT3 [Jatropha curcas] |
| 15 |
Hb_001801_020 |
0.1578618733 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 16 |
Hb_008014_040 |
0.1584508825 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 17 |
Hb_005649_080 |
0.1589058811 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 18 |
Hb_000037_220 |
0.1595190461 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 19 |
Hb_000085_260 |
0.1595427612 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_012738_030 |
0.1596133272 |
- |
- |
conserved hypothetical protein [Ricinus communis] |