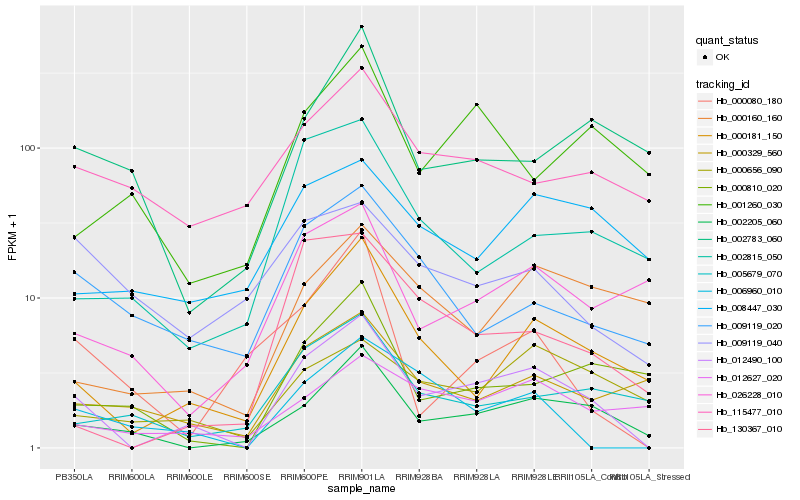
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012490_100 |
0.0 |
- |
- |
- |
| 2 |
Hb_000181_150 |
0.2251428104 |
- |
- |
hypothetical protein PHAVU_011G1647000g, partial [Phaseolus vulgaris] |
| 3 |
Hb_002205_060 |
0.2398510455 |
- |
- |
- |
| 4 |
Hb_006960_010 |
0.2645118719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_130367_010 |
0.2695313531 |
- |
- |
PREDICTED: sterol 3-beta-glucosyltransferase UGT80B1 [Jatropha curcas] |
| 6 |
Hb_000656_090 |
0.2812728083 |
- |
- |
PREDICTED: phosphoinositide phospholipase C 4-like [Jatropha curcas] |
| 7 |
Hb_009119_020 |
0.2812765451 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 8 |
Hb_000329_560 |
0.2831390111 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_012627_020 |
0.2844296901 |
- |
- |
PHYTOCHROME AND FLOWERING TIME 1 family protein [Populus trichocarpa] |
| 10 |
Hb_002815_050 |
0.285788391 |
- |
- |
PREDICTED: succinate-semialdehyde dehydrogenase, mitochondrial [Populus euphratica] |
| 11 |
Hb_115477_010 |
0.2884896845 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 12 |
Hb_005679_070 |
0.2887214016 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 13 |
Hb_008447_030 |
0.2909248558 |
- |
- |
- |
| 14 |
Hb_026228_010 |
0.2911401192 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000160_160 |
0.2936689285 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000810_020 |
0.2945098872 |
- |
- |
- |
| 17 |
Hb_000080_180 |
0.301491434 |
- |
- |
- |
| 18 |
Hb_001260_030 |
0.3031396607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009119_040 |
0.3034544545 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 20 |
Hb_002783_060 |
0.3062336533 |
- |
- |
PREDICTED: uncharacterized protein LOC105635325 [Jatropha curcas] |