| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
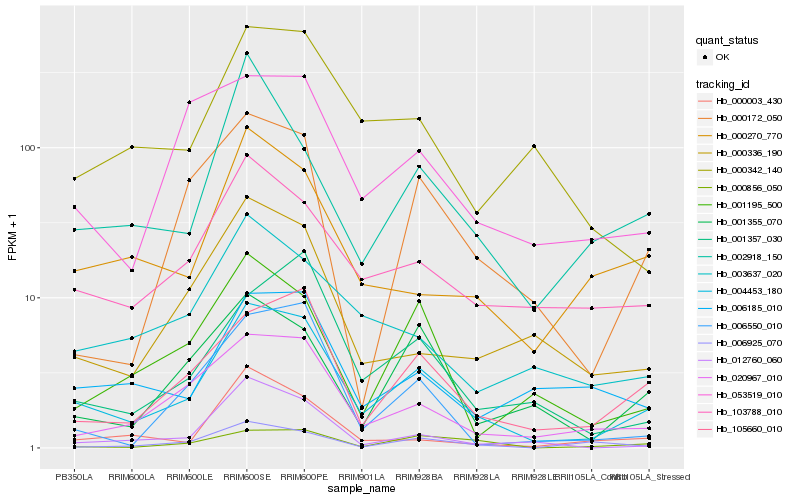
Hb_004453_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 2 |
Hb_105660_010 |
0.1639224384 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 3 |
Hb_020967_010 |
0.1669884189 |
- |
- |
- |
| 4 |
Hb_103788_010 |
0.1787973135 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 5 |
Hb_012760_060 |
0.1948284474 |
- |
- |
PREDICTED: uncharacterized protein LOC105647085 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003637_020 |
0.2007209895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000270_770 |
0.2016032697 |
- |
- |
PREDICTED: uncharacterized protein LOC105640410 [Jatropha curcas] |
| 8 |
Hb_006185_010 |
0.2023358841 |
- |
- |
hypothetical protein L484_013453 [Morus notabilis] |
| 9 |
Hb_006550_010 |
0.2040909623 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 10 |
Hb_000342_140 |
0.2044034907 |
- |
- |
PREDICTED: vacuolar-processing enzyme-like [Jatropha curcas] |
| 11 |
Hb_000336_190 |
0.2046231904 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Gossypium raimondii] |
| 12 |
Hb_001355_070 |
0.2056648269 |
- |
- |
P-loop containing nucleoside triphosphate hydrolases superfamily protein, putative isoform 2 [Theobroma cacao] |
| 13 |
Hb_001357_030 |
0.2072092595 |
- |
- |
hypothetical protein F383_04880 [Gossypium arboreum] |
| 14 |
Hb_001195_500 |
0.2089020684 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 15 |
Hb_053519_010 |
0.2093942904 |
- |
- |
PREDICTED: probable zinc transporter protein DDB_G0282067 [Jatropha curcas] |
| 16 |
Hb_000172_050 |
0.2100358113 |
- |
- |
PREDICTED: uncharacterized protein LOC105650611 [Jatropha curcas] |
| 17 |
Hb_000003_430 |
0.215501765 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 18 |
Hb_000856_050 |
0.2157559263 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 19 |
Hb_002918_150 |
0.2166743697 |
transcription factor |
TF Family: ERF |
ethylene-responsive element binding protein 1 [Hevea brasiliensis] |
| 20 |
Hb_006925_070 |
0.217367521 |
- |
- |
hypothetical protein JCGZ_20652 [Jatropha curcas] |