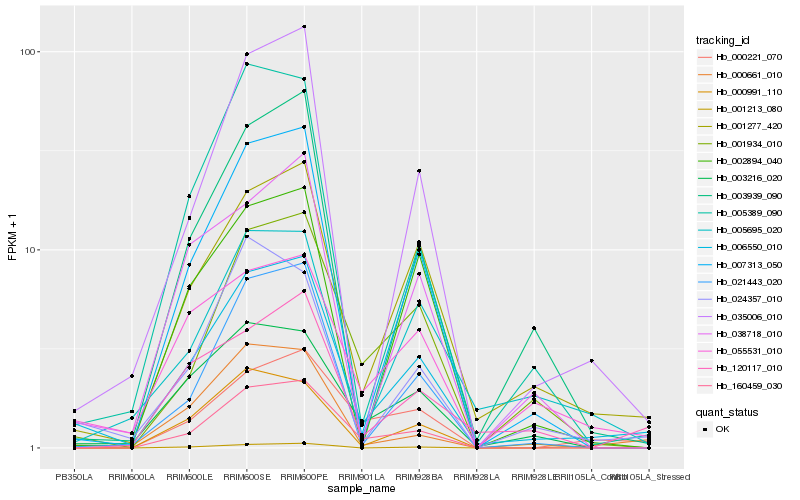
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006550_010 |
0.0 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 2 |
Hb_001277_420 |
0.1132294596 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 3 |
Hb_007313_050 |
0.1452936373 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 4 |
Hb_035006_010 |
0.151103185 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 5 |
Hb_024357_010 |
0.1599984584 |
- |
- |
PREDICTED: putative disease resistance protein RGA4, partial [Jatropha curcas] |
| 6 |
Hb_055531_010 |
0.1619162197 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 isoform X2 [Pyrus x bretschneideri] |
| 7 |
Hb_021443_020 |
0.1636653447 |
- |
- |
PREDICTED: uncharacterized protein LOC105648569 [Jatropha curcas] |
| 8 |
Hb_160459_030 |
0.1641878582 |
- |
- |
PREDICTED: uncharacterized protein LOC105648122 [Jatropha curcas] |
| 9 |
Hb_003939_090 |
0.1642512275 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_003216_020 |
0.1654409875 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 11 |
Hb_000661_010 |
0.1664080587 |
- |
- |
peroxisomal 3-keto-acyl-CoA thiolase [Hevea brasiliensis] |
| 12 |
Hb_005389_090 |
0.1706143143 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA27, putative [Ricinus communis] |
| 13 |
Hb_001934_010 |
0.1710658341 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_005695_020 |
0.1750237088 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 15 |
Hb_038718_010 |
0.1755679438 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 16 |
Hb_001213_080 |
0.175670907 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 17 |
Hb_120117_010 |
0.1789575027 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 18 |
Hb_000991_110 |
0.1795688172 |
- |
- |
PREDICTED: uncharacterized protein LOC105636336 [Jatropha curcas] |
| 19 |
Hb_002894_040 |
0.1801938282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000221_070 |
0.1806539484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |