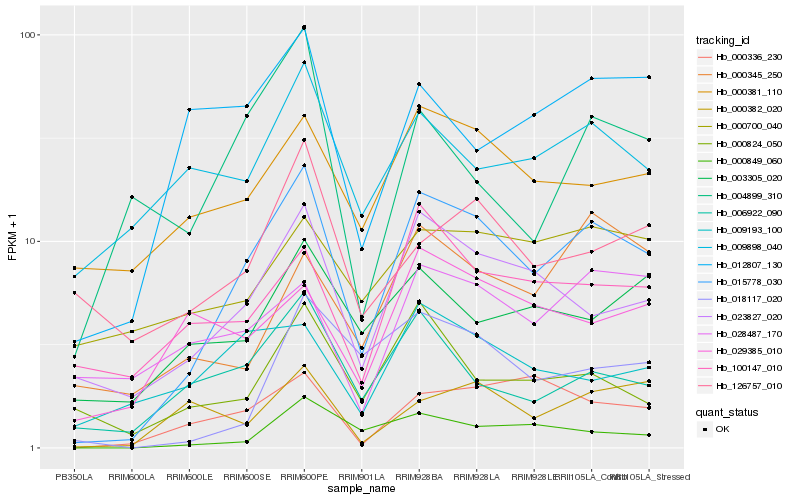
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015778_030 |
0.0 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 2 |
Hb_023827_020 |
0.175175755 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 3 |
Hb_000849_060 |
0.2012702739 |
- |
- |
putative sodium-dependent transporter yocS [Morus notabilis] |
| 4 |
Hb_000336_230 |
0.2035325039 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa007609mg [Prunus persica] |
| 5 |
Hb_006922_090 |
0.2137519765 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 6 |
Hb_018117_020 |
0.2149578659 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 7 |
Hb_000381_110 |
0.2169166063 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_012807_130 |
0.2209258546 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 9 |
Hb_100147_010 |
0.2233043701 |
- |
- |
PREDICTED: exosome complex component MTR3 [Jatropha curcas] |
| 10 |
Hb_003305_020 |
0.2281172135 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 11 |
Hb_000824_050 |
0.2288405423 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_000382_020 |
0.2294887598 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 13 |
Hb_009898_040 |
0.2328160355 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009193_100 |
0.2329374251 |
- |
- |
hypothetical protein JCGZ_15709 [Jatropha curcas] |
| 15 |
Hb_000345_250 |
0.2337902608 |
- |
- |
cullin 1-like protein [Hevea brasiliensis] |
| 16 |
Hb_028487_170 |
0.235501679 |
- |
- |
PREDICTED: uncharacterized protein LOC105634088 [Jatropha curcas] |
| 17 |
Hb_029385_010 |
0.2358496522 |
- |
- |
CTP synthase family protein [Populus trichocarpa] |
| 18 |
Hb_126757_010 |
0.2361360681 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_004899_310 |
0.2376208572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000700_040 |
0.238993158 |
- |
- |
UDP-Glycosyltransferase superfamily protein isoform 4 [Theobroma cacao] |