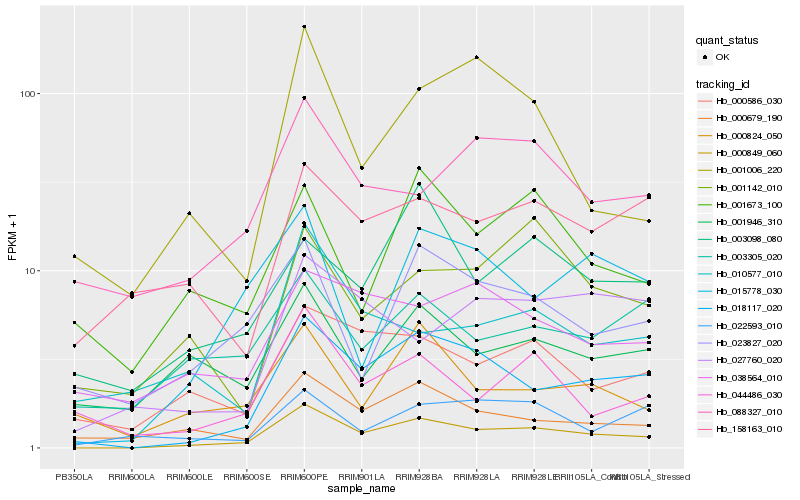
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000849_060 |
0.0 |
- |
- |
putative sodium-dependent transporter yocS [Morus notabilis] |
| 2 |
Hb_018117_020 |
0.1336801903 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 3 |
Hb_000679_190 |
0.1934451477 |
- |
- |
PREDICTED: uncharacterized protein LOC105637641 isoform X1 [Jatropha curcas] |
| 4 |
Hb_015778_030 |
0.2012702739 |
- |
- |
PREDICTED: probable myosin light chain kinase DDB_G0279831 [Jatropha curcas] |
| 5 |
Hb_000586_030 |
0.20339548 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 6 |
Hb_044486_030 |
0.2085999677 |
- |
- |
DMI1, partial [Cycas revoluta] |
| 7 |
Hb_023827_020 |
0.2110699749 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 8 |
Hb_088327_010 |
0.2188426401 |
- |
- |
PREDICTED: serine hydroxymethyltransferase 7 [Jatropha curcas] |
| 9 |
Hb_000824_050 |
0.2201804298 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 10 |
Hb_001006_220 |
0.2261622323 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 11 |
Hb_038564_010 |
0.2264703079 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 5 isoform X2 [Jatropha curcas] |
| 12 |
Hb_003098_080 |
0.227342572 |
- |
- |
- |
| 13 |
Hb_001946_310 |
0.2278265062 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 14 |
Hb_010577_010 |
0.2283704254 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 15 |
Hb_003305_020 |
0.2286564143 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 13-like [Jatropha curcas] |
| 16 |
Hb_001673_100 |
0.2291930507 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 17 |
Hb_158163_010 |
0.231085115 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_022593_010 |
0.2344183094 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001142_010 |
0.2369020919 |
- |
- |
PREDICTED: alcohol dehydrogenase-like 6 [Jatropha curcas] |
| 20 |
Hb_027760_020 |
0.2372893767 |
- |
- |
hypothetical protein M569_04462, partial [Genlisea aurea] |