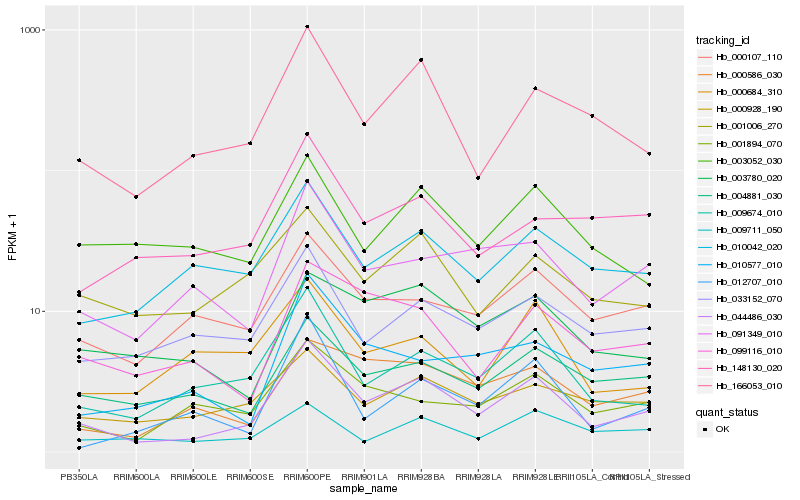
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044486_030 |
0.0 |
- |
- |
DMI1, partial [Cycas revoluta] |
| 2 |
Hb_009674_010 |
0.1410205377 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 3 |
Hb_166053_010 |
0.144698793 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 4 |
Hb_004881_030 |
0.16404033 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 5 |
Hb_091349_010 |
0.1653218625 |
- |
- |
hypothetical protein EUGRSUZ_C02287 [Eucalyptus grandis] |
| 6 |
Hb_010042_020 |
0.1696195981 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 7 |
Hb_033152_070 |
0.1714715327 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 8 |
Hb_000107_110 |
0.1717236049 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_000928_190 |
0.1723831092 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001894_070 |
0.1728933562 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001006_270 |
0.1747118932 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 12 |
Hb_000586_030 |
0.1770918176 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 13 |
Hb_099116_010 |
0.1796714174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010577_010 |
0.184160653 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase-like [Populus euphratica] |
| 15 |
Hb_012707_010 |
0.1882902574 |
- |
- |
PREDICTED: uncharacterized protein C594.04c [Jatropha curcas] |
| 16 |
Hb_009711_050 |
0.1884556612 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 17 |
Hb_003780_020 |
0.1894677999 |
- |
- |
PREDICTED: prolyl 4-hydroxylase subunit alpha-1 isoform X2 [Glycine max] |
| 18 |
Hb_148130_020 |
0.1900305577 |
- |
- |
- |
| 19 |
Hb_000684_310 |
0.1900363641 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g67720 [Jatropha curcas] |
| 20 |
Hb_003052_030 |
0.1917500073 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |