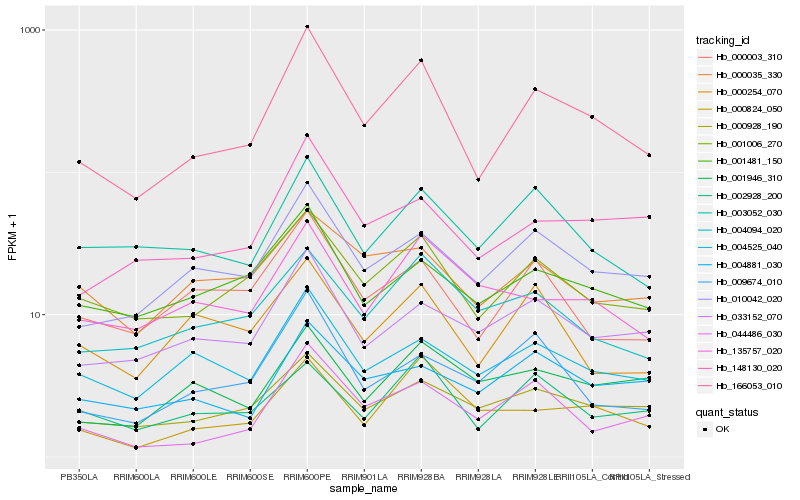
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_166053_010 |
0.0 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 2 |
Hb_001006_270 |
0.1195432976 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 3 |
Hb_010042_020 |
0.1261236867 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 4 |
Hb_000928_190 |
0.1408752646 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 5 |
Hb_148130_020 |
0.1445140995 |
- |
- |
- |
| 6 |
Hb_044486_030 |
0.144698793 |
- |
- |
DMI1, partial [Cycas revoluta] |
| 7 |
Hb_000003_310 |
0.1471389035 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 8 |
Hb_004525_040 |
0.147777855 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 9 |
Hb_033152_070 |
0.1481931165 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 10 |
Hb_000824_050 |
0.150805329 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_003052_030 |
0.1516004782 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_001481_150 |
0.1527015144 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 13 |
Hb_002928_200 |
0.1562267563 |
- |
- |
PREDICTED: Niemann-Pick C1 protein-like [Jatropha curcas] |
| 14 |
Hb_009674_010 |
0.1566586594 |
- |
- |
AAEL007687-PA [Aedes aegypti] |
| 15 |
Hb_004094_020 |
0.1566750265 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_004881_030 |
0.1634711278 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001946_310 |
0.1641833019 |
- |
- |
PREDICTED: probable ethanolamine kinase isoform X2 [Jatropha curcas] |
| 18 |
Hb_135757_020 |
0.1649369911 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 19 |
Hb_000254_070 |
0.165501804 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 20 |
Hb_000035_330 |
0.1686489648 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |