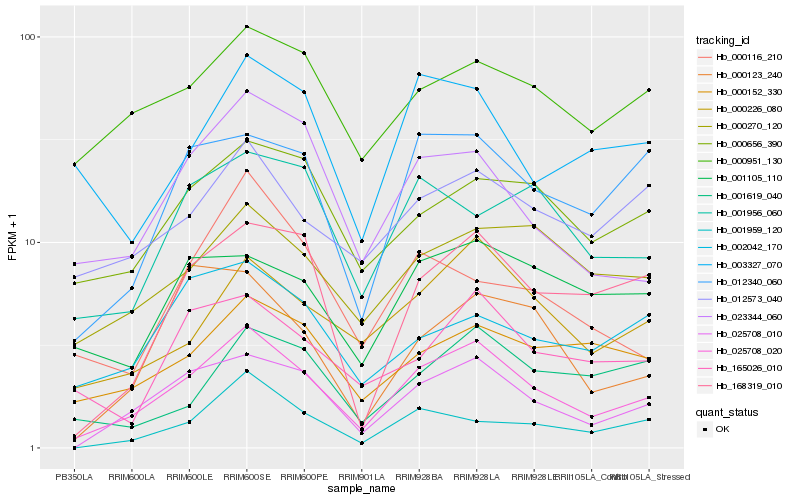
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_025708_020 |
0.0 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 2 |
Hb_025708_010 |
0.1100679422 |
- |
- |
hypothetical protein MTR_1g051060 [Medicago truncatula] |
| 3 |
Hb_000226_080 |
0.1473879976 |
- |
- |
prephenate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_012340_060 |
0.1525753269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_168319_010 |
0.1566777941 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_023344_060 |
0.1592318633 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA11 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000656_390 |
0.1649631356 |
- |
- |
PREDICTED: probable GTP diphosphokinase RSH2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000270_120 |
0.1666652844 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g18900 [Jatropha curcas] |
| 9 |
Hb_000152_330 |
0.1667272076 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 6-like [Jatropha curcas] |
| 10 |
Hb_165026_010 |
0.1685421446 |
transcription factor |
TF Family: NAC |
hypothetical protein JCGZ_14166 [Jatropha curcas] |
| 11 |
Hb_000951_130 |
0.1687935774 |
- |
- |
PREDICTED: transcription factor GTE2-like [Jatropha curcas] |
| 12 |
Hb_012573_040 |
0.1705196734 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g55270-like [Jatropha curcas] |
| 13 |
Hb_001959_120 |
0.1745315859 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 14 |
Hb_001956_060 |
0.1769463693 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 3C-like [Jatropha curcas] |
| 15 |
Hb_000123_240 |
0.1774400736 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 16 |
Hb_001105_110 |
0.1775665707 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002042_170 |
0.1807431437 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 18 |
Hb_003327_070 |
0.1819052587 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 19 |
Hb_001619_040 |
0.1820079372 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 20 |
Hb_000116_210 |
0.1840184974 |
- |
- |
PREDICTED: uncharacterized protein LOC105628567 [Jatropha curcas] |