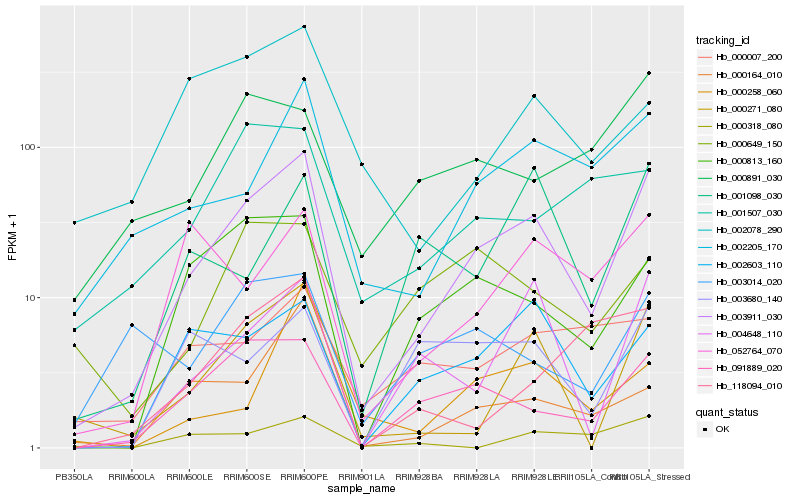
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003911_030 |
0.0 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_000271_080 |
0.2070467307 |
- |
- |
hypothetical protein POPTR_0006s08460g [Populus trichocarpa] |
| 3 |
Hb_091889_020 |
0.2102404813 |
- |
- |
- |
| 4 |
Hb_002205_170 |
0.2111839701 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 5 |
Hb_004648_110 |
0.224972739 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_000813_160 |
0.2330125921 |
- |
- |
hypothetical protein PRUPE_ppa013181mg [Prunus persica] |
| 7 |
Hb_002603_110 |
0.2339180817 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 8 |
Hb_000164_010 |
0.2591871962 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 9 |
Hb_052764_070 |
0.2608381996 |
- |
- |
PREDICTED: uncharacterized protein LOC105649473 [Jatropha curcas] |
| 10 |
Hb_000891_030 |
0.2623717535 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 11 |
Hb_001098_030 |
0.2626716764 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Populus euphratica] |
| 12 |
Hb_001507_030 |
0.2666957642 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 2 [Jatropha curcas] |
| 13 |
Hb_003680_140 |
0.2677996122 |
- |
- |
PREDICTED: uncharacterized protein LOC105121770 [Populus euphratica] |
| 14 |
Hb_000007_200 |
0.2708205399 |
- |
- |
PREDICTED: choline/ethanolaminephosphotransferase 1 [Jatropha curcas] |
| 15 |
Hb_002078_290 |
0.2725929375 |
- |
- |
PREDICTED: uncharacterized protein LOC100852520 [Vitis vinifera] |
| 16 |
Hb_000318_080 |
0.2736642274 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 17 |
Hb_118094_010 |
0.2739086764 |
- |
- |
hypothetical protein POPTR_0011s06595g [Populus trichocarpa] |
| 18 |
Hb_003014_020 |
0.2739697114 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 19 |
Hb_000649_150 |
0.2755959961 |
- |
- |
PREDICTED: uncharacterized protein LOC105634115 [Jatropha curcas] |
| 20 |
Hb_000258_060 |
0.2780987998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |