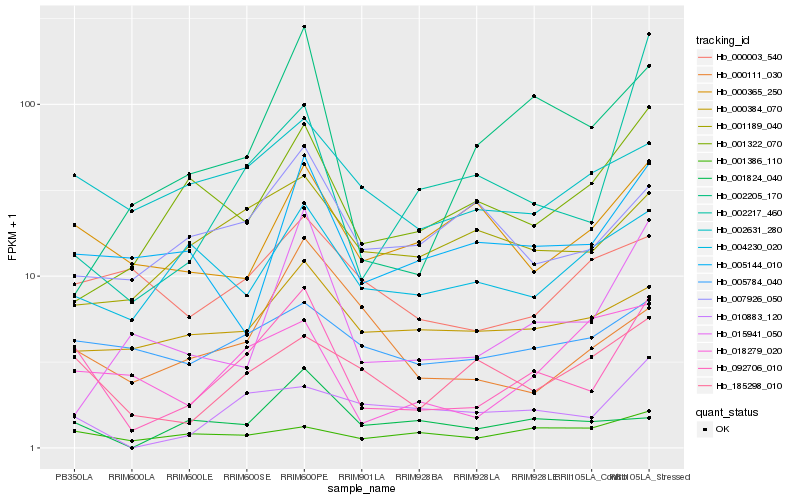
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_092706_010 |
0.0 |
- |
- |
PREDICTED: beta-galactosidase-like [Jatropha curcas] |
| 2 |
Hb_185298_010 |
0.2453718163 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 3 |
Hb_010883_120 |
0.2493857631 |
- |
- |
- |
| 4 |
Hb_002631_280 |
0.2500176752 |
- |
- |
PREDICTED: ubiquitin isoform X2 [Eucalyptus grandis] |
| 5 |
Hb_005144_010 |
0.2530458695 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 6 |
Hb_018279_020 |
0.2566946587 |
- |
- |
hypothetical protein JCGZ_03682 [Jatropha curcas] |
| 7 |
Hb_000365_250 |
0.2602572421 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000111_030 |
0.2602964119 |
- |
- |
PREDICTED: uncharacterized protein LOC104094030 [Nicotiana tomentosiformis] |
| 9 |
Hb_005784_040 |
0.2624165499 |
- |
- |
PREDICTED: uncharacterized protein LOC105645160 isoform X1 [Jatropha curcas] |
| 10 |
Hb_015941_050 |
0.2649844218 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002217_460 |
0.267803333 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 12 |
Hb_000384_070 |
0.2682544266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007926_050 |
0.2688021447 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 14 |
Hb_001824_040 |
0.2694489305 |
- |
- |
hypothetical protein M569_11359, partial [Genlisea aurea] |
| 15 |
Hb_001386_110 |
0.2695103662 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 16 |
Hb_000003_540 |
0.2707123022 |
- |
- |
PREDICTED: homeobox-leucine zipper protein ATHB-15 [Jatropha curcas] |
| 17 |
Hb_002205_170 |
0.2709863385 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 18 |
Hb_004230_020 |
0.2710384773 |
- |
- |
PREDICTED: uncharacterized protein LOC105648105 [Jatropha curcas] |
| 19 |
Hb_001189_040 |
0.2730154271 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 20 |
Hb_001322_070 |
0.273970907 |
- |
- |
protein with unknown function [Ricinus communis] |