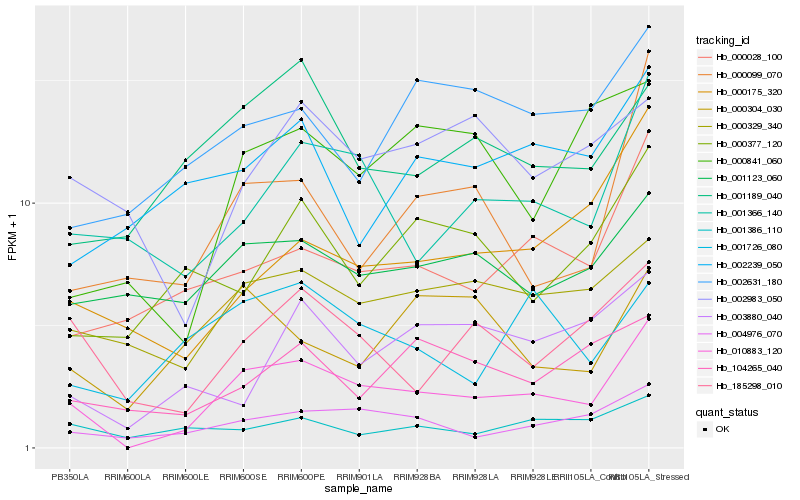
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010883_120 |
0.0 |
- |
- |
- |
| 2 |
Hb_004976_070 |
0.1901741437 |
- |
- |
eukaryotic translation initiation factor 2b, epsilon subunit, putative [Ricinus communis] |
| 3 |
Hb_000329_340 |
0.2027643006 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001366_140 |
0.2040073254 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 5 |
Hb_185298_010 |
0.2054548874 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 6 |
Hb_000028_100 |
0.2103119308 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 7 |
Hb_001123_060 |
0.2207661532 |
- |
- |
PREDICTED: ALBINO3-like protein 2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000099_070 |
0.2220454472 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 9 |
Hb_000175_320 |
0.222417445 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 10 |
Hb_001189_040 |
0.2224716162 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 11 |
Hb_001726_080 |
0.2252122141 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000377_120 |
0.2314002865 |
- |
- |
PREDICTED: acyl carrier protein 3, mitochondrial isoform X2 [Jatropha curcas] |
| 13 |
Hb_002631_180 |
0.2317625805 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-2 [Jatropha curcas] |
| 14 |
Hb_000841_060 |
0.2342179194 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0015s06470g [Populus trichocarpa] |
| 15 |
Hb_001386_110 |
0.2344147417 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 16 |
Hb_003880_040 |
0.2353978167 |
- |
- |
hypothetical protein POPTR_0001s30770g [Populus trichocarpa] |
| 17 |
Hb_104265_040 |
0.2355546718 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002983_050 |
0.236179532 |
- |
- |
PREDICTED: acetylornithine aminotransferase, mitochondrial [Jatropha curcas] |
| 19 |
Hb_002239_050 |
0.2368799527 |
- |
- |
PREDICTED: isovaleryl-CoA dehydrogenase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000304_030 |
0.2380694647 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |