| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
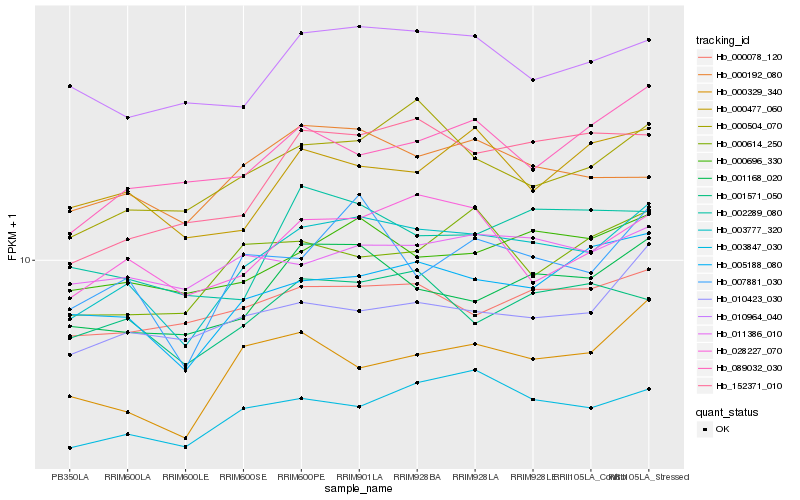
Hb_003777_320 |
0.0 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 15 homolog [Jatropha curcas] |
| 2 |
Hb_005188_080 |
0.081623625 |
- |
- |
Serine/threonine-protein kinase PBS1, putative [Ricinus communis] |
| 3 |
Hb_001168_020 |
0.0820509427 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit gamma [Jatropha curcas] |
| 4 |
Hb_003847_030 |
0.0832469143 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Jatropha curcas] |
| 5 |
Hb_007881_030 |
0.0842997222 |
- |
- |
Protein YME1, putative [Ricinus communis] |
| 6 |
Hb_000696_330 |
0.0844974397 |
- |
- |
PREDICTED: uncharacterized protein LOC105647254 [Jatropha curcas] |
| 7 |
Hb_000504_070 |
0.0886504832 |
- |
- |
PREDICTED: pescadillo homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_152371_010 |
0.0915734085 |
- |
- |
RNA-binding KH domain-containing protein [Theobroma cacao] |
| 9 |
Hb_028227_070 |
0.091736129 |
- |
- |
PREDICTED: uncharacterized protein LOC105644820 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000329_340 |
0.0920775153 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010964_040 |
0.0947424887 |
- |
- |
Eukaryotic initiation factor 4A-3 [Gossypium arboreum] |
| 12 |
Hb_000078_120 |
0.0954747539 |
- |
- |
PREDICTED: BRCA1-associated protein [Jatropha curcas] |
| 13 |
Hb_001571_050 |
0.0960768921 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 14 |
Hb_010423_030 |
0.096100544 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |
| 15 |
Hb_011386_010 |
0.0975202358 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 16 |
Hb_000192_080 |
0.0978323699 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_089032_030 |
0.0992512828 |
- |
- |
PREDICTED: 3-isopropylmalate dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000614_250 |
0.0995284698 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 19 |
Hb_000477_060 |
0.1008557698 |
- |
- |
PREDICTED: serine-threonine kinase receptor-associated protein-like [Jatropha curcas] |
| 20 |
Hb_002289_080 |
0.1021949295 |
- |
- |
hypothetical protein POPTR_0016s05590g [Populus trichocarpa] |