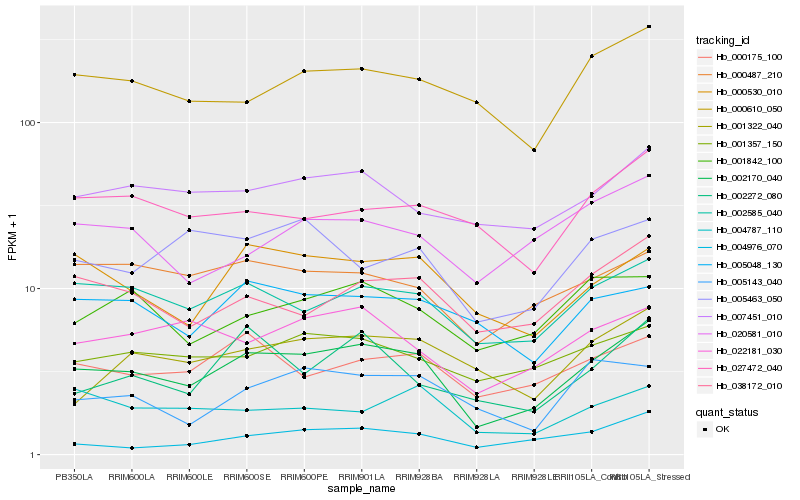
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002170_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002585_040 |
0.1166284232 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 3 |
Hb_000530_010 |
0.1301085162 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03 [Jatropha curcas] |
| 4 |
Hb_038172_010 |
0.1305438051 |
desease resistance |
Gene Name: AAA |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 5 |
Hb_000175_100 |
0.135104588 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_022181_030 |
0.1367075563 |
- |
- |
PREDICTED: uncharacterized protein LOC105649615 isoform X2 [Jatropha curcas] |
| 7 |
Hb_007451_010 |
0.1369020748 |
- |
- |
PREDICTED: 60S ribosome subunit biogenesis protein NIP7 homolog [Jatropha curcas] |
| 8 |
Hb_000487_210 |
0.1378372875 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |
| 9 |
Hb_000610_050 |
0.1388755101 |
- |
- |
skp1, putative [Ricinus communis] |
| 10 |
Hb_001842_100 |
0.1404211528 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
hypothetical protein JCGZ_10905 [Jatropha curcas] |
| 11 |
Hb_001357_150 |
0.1404560473 |
- |
- |
hypothetical protein POPTR_0011s08930g [Populus trichocarpa] |
| 12 |
Hb_005048_130 |
0.1467604114 |
- |
- |
hypothetical protein POPTR_0004s03700g [Populus trichocarpa] |
| 13 |
Hb_001322_040 |
0.148130078 |
- |
- |
- |
| 14 |
Hb_020581_010 |
0.148530659 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_002272_080 |
0.1487424697 |
- |
- |
- |
| 16 |
Hb_004976_070 |
0.1490187617 |
- |
- |
eukaryotic translation initiation factor 2b, epsilon subunit, putative [Ricinus communis] |
| 17 |
Hb_004787_110 |
0.1504980516 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_005143_040 |
0.150910219 |
- |
- |
hypothetical protein JCGZ_17315 [Jatropha curcas] |
| 19 |
Hb_005463_050 |
0.1510056082 |
- |
- |
PREDICTED: uncharacterized protein LOC105632514 [Jatropha curcas] |
| 20 |
Hb_027472_040 |
0.1510838784 |
- |
- |
SKP1 [Hevea brasiliensis] |