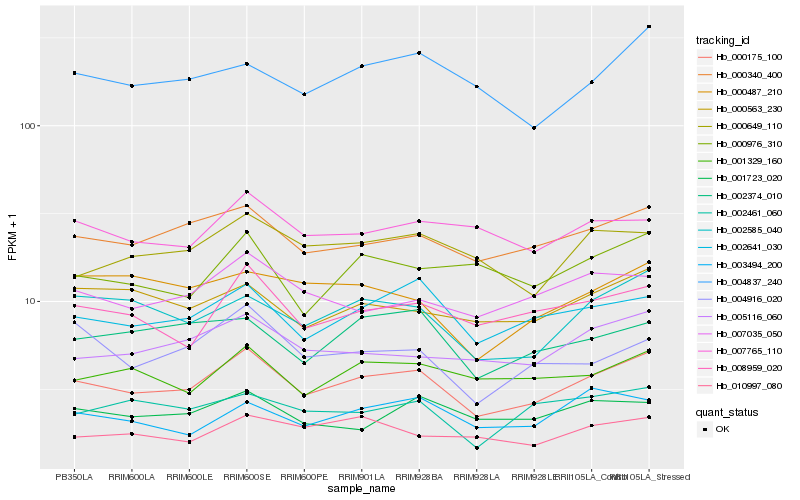
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000175_100 |
0.0 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 2 |
Hb_000340_400 |
0.0794413095 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 3 |
Hb_002641_030 |
0.0815926681 |
- |
- |
PREDICTED: flowering time control protein FCA isoform X1 [Jatropha curcas] |
| 4 |
Hb_002585_040 |
0.0850488453 |
- |
- |
PREDICTED: cell division control protein 48 homolog B [Jatropha curcas] |
| 5 |
Hb_008959_020 |
0.0886978264 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 6 |
Hb_005116_060 |
0.0912010908 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001329_160 |
0.0956940865 |
- |
- |
PREDICTED: uncharacterized protein LOC105642679 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004837_240 |
0.0978902799 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |
| 9 |
Hb_000563_230 |
0.0984233728 |
- |
- |
phospholipase d zeta, putative [Ricinus communis] |
| 10 |
Hb_007035_050 |
0.1002383451 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 11 |
Hb_002374_010 |
0.1002629405 |
- |
- |
PREDICTED: protein translocase subunit SECA2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000649_110 |
0.1026488576 |
- |
- |
PREDICTED: protein VAC14 homolog [Jatropha curcas] |
| 13 |
Hb_004916_020 |
0.1043299147 |
- |
- |
PREDICTED: DNA ligase 4 [Jatropha curcas] |
| 14 |
Hb_002461_060 |
0.104432662 |
- |
- |
PREDICTED: SWR1-complex protein 4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_007765_110 |
0.1047213048 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 40-like [Jatropha curcas] |
| 16 |
Hb_000976_310 |
0.1055551247 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_010997_080 |
0.1056723277 |
transcription factor |
TF Family: mTERF |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_001723_020 |
0.1058986183 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 19 |
Hb_003494_200 |
0.1061013605 |
- |
- |
PREDICTED: beta-1,3-galactosyltransferase 15-like [Jatropha curcas] |
| 20 |
Hb_000487_210 |
0.1076720272 |
- |
- |
PREDICTED: uncharacterized protein LOC105641929 [Jatropha curcas] |