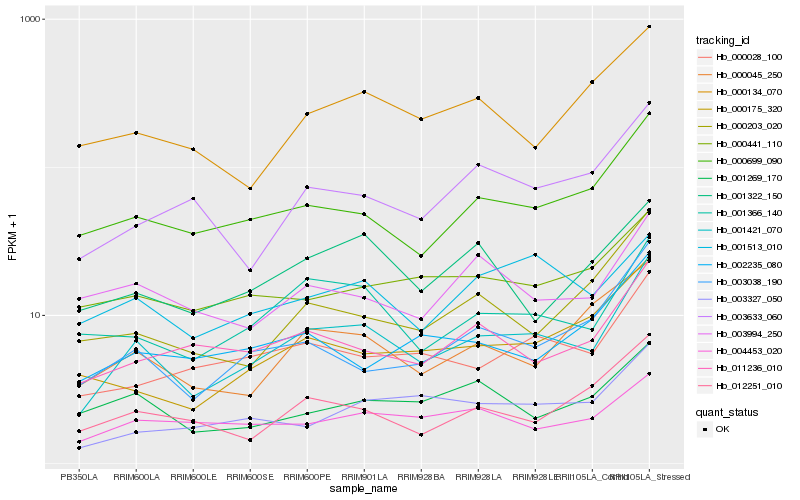
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001421_070 |
0.0 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 2 |
Hb_001366_140 |
0.1350526437 |
- |
- |
hypothetical protein POPTR_0014s10420g [Populus trichocarpa] |
| 3 |
Hb_000699_090 |
0.1361277781 |
- |
- |
cyclophilin, putative [Ricinus communis] |
| 4 |
Hb_000028_100 |
0.1424472546 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 5 |
Hb_003038_190 |
0.1471661346 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |
| 6 |
Hb_011236_010 |
0.1509126923 |
- |
- |
hypothetical protein JCGZ_17006 [Jatropha curcas] |
| 7 |
Hb_003327_050 |
0.1566317934 |
- |
- |
surfeit locus protein, putative [Ricinus communis] |
| 8 |
Hb_003633_060 |
0.1569033225 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 9 |
Hb_000045_250 |
0.1569106259 |
- |
- |
PREDICTED: uncharacterized protein LOC105646572 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000175_320 |
0.1613786964 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 11 |
Hb_004453_020 |
0.1616789624 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_012251_010 |
0.1621116625 |
- |
- |
- |
| 13 |
Hb_000203_020 |
0.1622586684 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 14 |
Hb_001513_010 |
0.1631048337 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000134_070 |
0.1671118935 |
- |
- |
60S ribosomal protein L6, putative [Ricinus communis] |
| 16 |
Hb_000441_110 |
0.1672844113 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 17 |
Hb_001322_150 |
0.1686593732 |
- |
- |
PREDICTED: vesicle-associated protein 2-1 [Jatropha curcas] |
| 18 |
Hb_002235_080 |
0.1695556593 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 19 |
Hb_003994_250 |
0.1696930167 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 20 |
Hb_001269_170 |
0.1697461462 |
- |
- |
PREDICTED: uncharacterized protein At2g39795, mitochondrial-like [Jatropha curcas] |