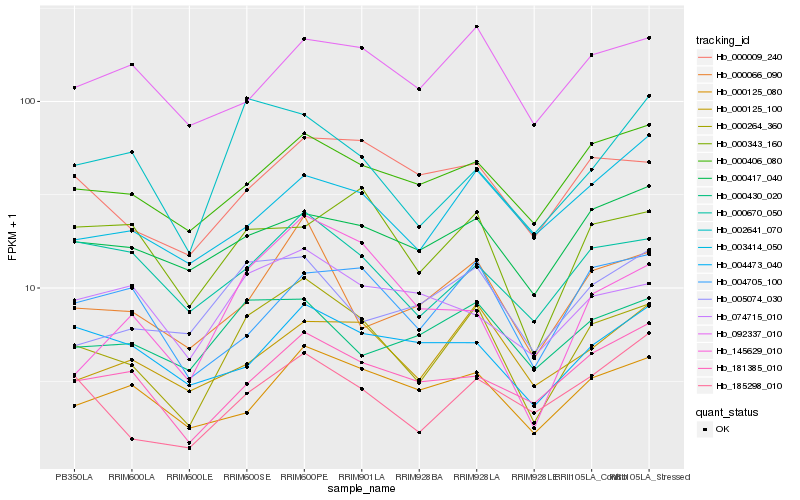
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000264_360 |
0.0 |
- |
- |
hypothetical protein JCGZ_21443 [Jatropha curcas] |
| 2 |
Hb_000066_090 |
0.1464721672 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme-like [Jatropha curcas] |
| 3 |
Hb_000406_080 |
0.1524980704 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 6B [Jatropha curcas] |
| 4 |
Hb_000125_080 |
0.1536406386 |
transcription factor |
TF Family: mTERF |
- |
| 5 |
Hb_000009_240 |
0.1549064599 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000670_050 |
0.1556956481 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |
| 7 |
Hb_181385_010 |
0.1574956938 |
- |
- |
PREDICTED: uncharacterized protein LOC105650319 [Jatropha curcas] |
| 8 |
Hb_000125_100 |
0.1591835758 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105635070 [Jatropha curcas] |
| 9 |
Hb_002641_070 |
0.1600070547 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 10 |
Hb_004705_100 |
0.1606407389 |
- |
- |
PREDICTED: inactive leucine-rich repeat receptor-like protein kinase CORYNE [Jatropha curcas] |
| 11 |
Hb_185298_010 |
0.1628564316 |
- |
- |
PREDICTED: biotin--protein ligase 2-like [Jatropha curcas] |
| 12 |
Hb_000343_160 |
0.1631933399 |
- |
- |
PREDICTED: protein SYS1 homolog [Jatropha curcas] |
| 13 |
Hb_005074_030 |
0.1645865204 |
- |
- |
- |
| 14 |
Hb_000430_020 |
0.1647078184 |
- |
- |
- |
| 15 |
Hb_000417_040 |
0.1663991079 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 16 |
Hb_092337_010 |
0.168364134 |
- |
- |
PREDICTED: actin-depolymerizing factor 2-like [Nelumbo nucifera] |
| 17 |
Hb_145629_010 |
0.1713706868 |
- |
- |
- |
| 18 |
Hb_074715_010 |
0.1719727369 |
- |
- |
PREDICTED: disease resistance protein RPS6-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_003414_050 |
0.1739330937 |
- |
- |
PREDICTED: uncharacterized protein LOC105640180 isoform X3 [Jatropha curcas] |
| 20 |
Hb_004473_040 |
0.1748385947 |
- |
- |
conserved hypothetical protein [Ricinus communis] |