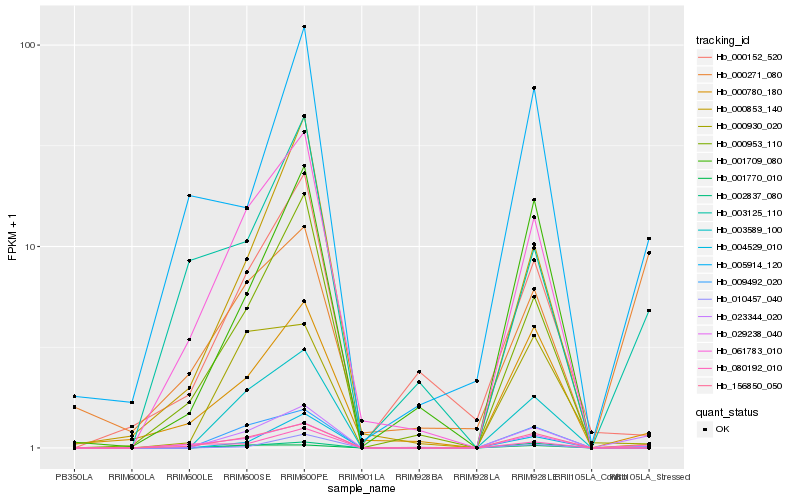
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023344_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_001770_010 |
0.1752151138 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 3 |
Hb_010457_040 |
0.2345734978 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 4 |
Hb_080192_010 |
0.2492452591 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 5 |
Hb_003589_100 |
0.2557736415 |
- |
- |
hypothetical protein JCGZ_03338 [Jatropha curcas] |
| 6 |
Hb_009492_020 |
0.2610930098 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 7 |
Hb_000271_080 |
0.2686817842 |
- |
- |
hypothetical protein POPTR_0006s08460g [Populus trichocarpa] |
| 8 |
Hb_000780_180 |
0.270196748 |
- |
- |
PREDICTED: uncharacterized protein LOC105641547 [Jatropha curcas] |
| 9 |
Hb_002837_080 |
0.2743547396 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 10 |
Hb_005914_120 |
0.2793164422 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 11 |
Hb_156850_050 |
0.2796633215 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |
| 12 |
Hb_004529_010 |
0.2808511673 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 13 |
Hb_003125_110 |
0.2864400052 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 14 |
Hb_000930_020 |
0.2907347832 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 15 |
Hb_000953_110 |
0.2908660798 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 16 |
Hb_061783_010 |
0.2925234483 |
- |
- |
PREDICTED: uncharacterized protein LOC105124983 isoform X4 [Populus euphratica] |
| 17 |
Hb_000853_140 |
0.2946057084 |
- |
- |
hypothetical protein POPTR_0014s12080g [Populus trichocarpa] |
| 18 |
Hb_029238_040 |
0.29964449 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 19 |
Hb_000152_520 |
0.3002726328 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 20 |
Hb_001709_080 |
0.3003837019 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |