| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
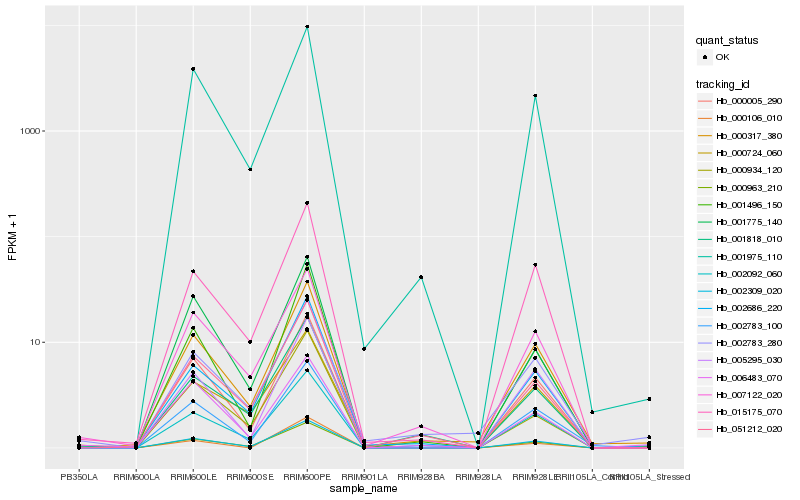
Hb_000963_210 |
0.0 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 2 |
Hb_002309_020 |
0.1224258332 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 3 |
Hb_000317_380 |
0.1260541496 |
- |
- |
Peroxidase 47 precursor, putative [Ricinus communis] |
| 4 |
Hb_002092_060 |
0.1300751035 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000005_290 |
0.132200008 |
- |
- |
Serine carboxypeptidase, putative [Ricinus communis] |
| 6 |
Hb_002686_220 |
0.1331433575 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 7 |
Hb_001975_110 |
0.1360482339 |
- |
- |
- |
| 8 |
Hb_015175_070 |
0.1428092756 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 9 |
Hb_051212_020 |
0.1454999511 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 10 |
Hb_002783_280 |
0.1463588243 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 11 |
Hb_006483_070 |
0.1486282314 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 12 |
Hb_000724_060 |
0.1493773065 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 13 |
Hb_000934_120 |
0.1501288532 |
- |
- |
PREDICTED: tumor necrosis factor ligand superfamily member 6 [Populus euphratica] |
| 14 |
Hb_001775_140 |
0.1504064204 |
- |
- |
Phytosulfokine 3 precursor, putative isoform 2 [Theobroma cacao] |
| 15 |
Hb_001496_150 |
0.1516585877 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 16 |
Hb_000106_010 |
0.1524081038 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 17 |
Hb_002783_100 |
0.1526085183 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 18 |
Hb_005295_030 |
0.1535658028 |
- |
- |
PREDICTED: uncharacterized protein LOC105645488 [Jatropha curcas] |
| 19 |
Hb_007122_020 |
0.1536953161 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 20 |
Hb_001818_010 |
0.1566721032 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |