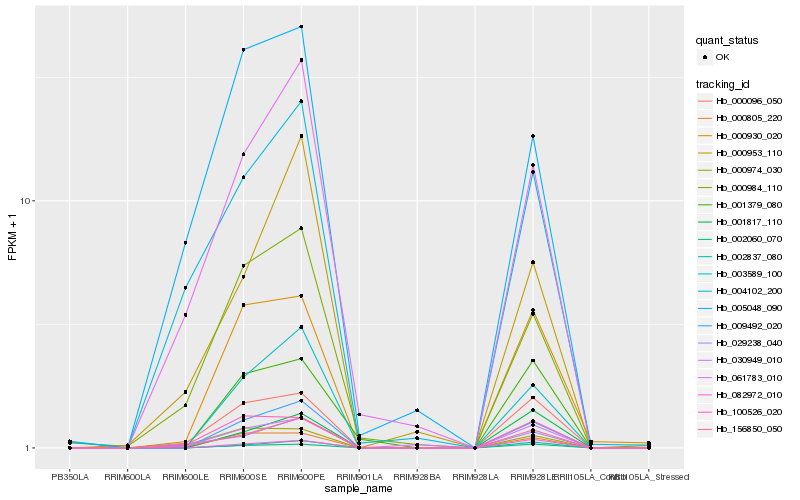
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009492_020 |
0.0 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 2 |
Hb_003589_100 |
0.0474668268 |
- |
- |
hypothetical protein JCGZ_03338 [Jatropha curcas] |
| 3 |
Hb_000805_220 |
0.1122639703 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_002837_080 |
0.1218755875 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002060_070 |
0.1307251613 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 6 |
Hb_156850_050 |
0.1359601849 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |
| 7 |
Hb_000930_020 |
0.145820817 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 8 |
Hb_000096_050 |
0.1531734862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001379_080 |
0.1608242987 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Vitis vinifera] |
| 10 |
Hb_082972_010 |
0.1652525317 |
- |
- |
- |
| 11 |
Hb_061783_010 |
0.1656385534 |
- |
- |
PREDICTED: uncharacterized protein LOC105124983 isoform X4 [Populus euphratica] |
| 12 |
Hb_000984_110 |
0.1745868625 |
- |
- |
hypothetical protein JCGZ_15953 [Jatropha curcas] |
| 13 |
Hb_001817_110 |
0.1774119181 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 14 |
Hb_029238_040 |
0.1857279285 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 15 |
Hb_100526_020 |
0.1858386616 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 16 |
Hb_030949_010 |
0.1868984642 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 17 |
Hb_004102_200 |
0.1936812958 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 18 |
Hb_000974_030 |
0.1973826933 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 19 |
Hb_000953_110 |
0.2011421732 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105629453 [Jatropha curcas] |
| 20 |
Hb_005048_090 |
0.2042921555 |
- |
- |
PREDICTED: glycine-rich cell wall structural protein 2 [Jatropha curcas] |