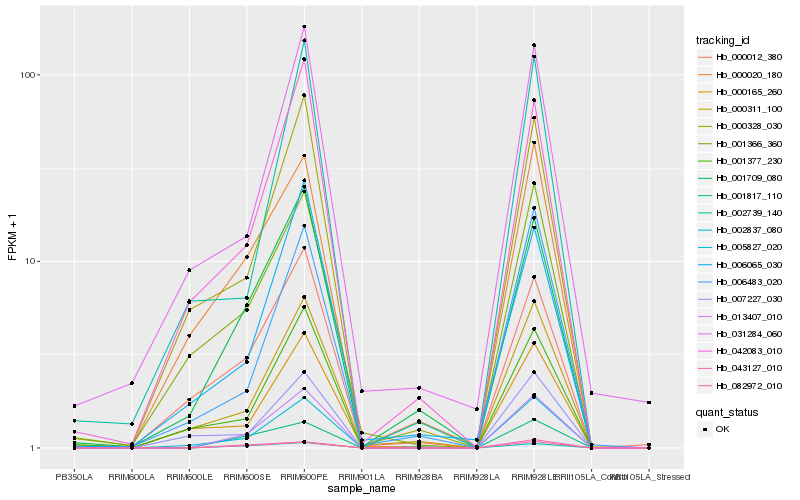
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013407_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_23575 [Jatropha curcas] |
| 2 |
Hb_005827_020 |
0.115423042 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 3 |
Hb_001817_110 |
0.1176026738 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 4 |
Hb_002837_080 |
0.1190318433 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_043127_010 |
0.119820079 |
- |
- |
PREDICTED: uncharacterized protein LOC105648822 [Jatropha curcas] |
| 6 |
Hb_001709_080 |
0.1277546887 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_000311_100 |
0.1422831026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000328_030 |
0.1448650167 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |
| 9 |
Hb_082972_010 |
0.1462080592 |
- |
- |
- |
| 10 |
Hb_042083_010 |
0.1500916014 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 8 [UDP-forming] [Jatropha curcas] |
| 11 |
Hb_001377_230 |
0.1501339697 |
- |
- |
PREDICTED: uncharacterized protein LOC105135159 [Populus euphratica] |
| 12 |
Hb_000020_180 |
0.1565393943 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_006483_020 |
0.1602098125 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_001366_360 |
0.1611934881 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000012_380 |
0.1612434491 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006065_030 |
0.1621413414 |
- |
- |
PREDICTED: filament-like plant protein 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000165_260 |
0.1638066896 |
- |
- |
PREDICTED: uncharacterized protein LOC105133752 isoform X2 [Populus euphratica] |
| 18 |
Hb_007227_030 |
0.1642430753 |
- |
- |
hypothetical protein JCGZ_24975 [Jatropha curcas] |
| 19 |
Hb_002739_140 |
0.1666902681 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |
| 20 |
Hb_031284_060 |
0.1670670354 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 7 [UDP-forming] [Jatropha curcas] |