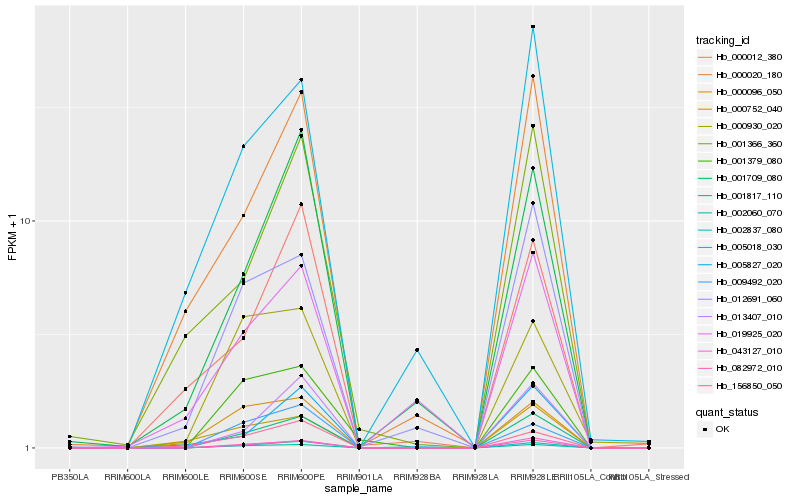
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001817_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 2 |
Hb_082972_010 |
0.0302547192 |
- |
- |
- |
| 3 |
Hb_043127_010 |
0.0537670402 |
- |
- |
PREDICTED: uncharacterized protein LOC105648822 [Jatropha curcas] |
| 4 |
Hb_002837_080 |
0.0651327771 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_002060_070 |
0.0876348429 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 6 |
Hb_013407_010 |
0.1176026738 |
- |
- |
hypothetical protein JCGZ_23575 [Jatropha curcas] |
| 7 |
Hb_012691_060 |
0.1444252589 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like isoform X1 [Prunus mume] |
| 8 |
Hb_000020_180 |
0.1565504632 |
- |
- |
laccase, putative [Ricinus communis] |
| 9 |
Hb_001379_080 |
0.1603292541 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Vitis vinifera] |
| 10 |
Hb_001709_080 |
0.1673876703 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 11 |
Hb_005018_030 |
0.168472234 |
- |
- |
hypothetical protein CICLE_v10019811mg [Citrus clementina] |
| 12 |
Hb_005827_020 |
0.1699401928 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 13 |
Hb_000096_050 |
0.1718600082 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_009492_020 |
0.1774119181 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 15 |
Hb_156850_050 |
0.1789105771 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |
| 16 |
Hb_000752_040 |
0.1823235501 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_001366_360 |
0.1841765143 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_019925_020 |
0.1857122912 |
- |
- |
electron carrier, putative [Ricinus communis] |
| 19 |
Hb_000930_020 |
0.1931781095 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 20 |
Hb_000012_380 |
0.197531602 |
transcription factor |
TF Family: MYB-related |
conserved hypothetical protein [Ricinus communis] |