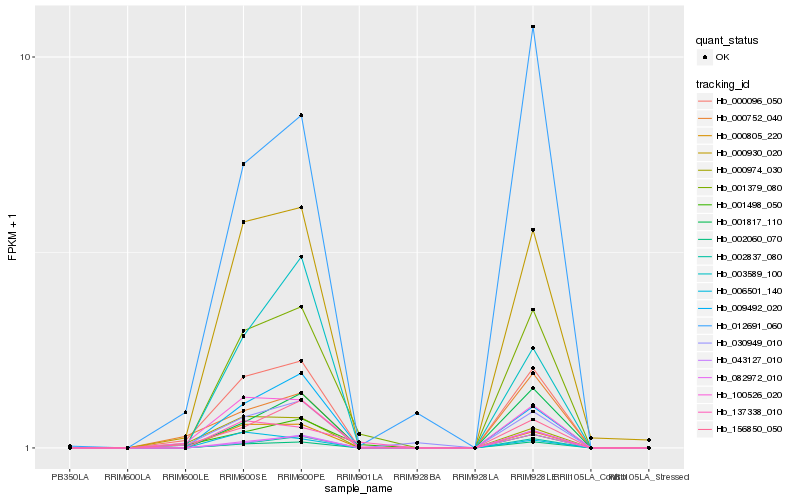
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001379_080 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Vitis vinifera] |
| 2 |
Hb_002060_070 |
0.1133829841 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 3 |
Hb_082972_010 |
0.14138649 |
- |
- |
- |
| 4 |
Hb_000805_220 |
0.1508124995 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 5 |
Hb_002837_080 |
0.1543983731 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000930_020 |
0.1551198982 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 7 |
Hb_100526_020 |
0.1553067373 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 8 |
Hb_000096_050 |
0.1559384562 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001817_110 |
0.1603292541 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 10 |
Hb_009492_020 |
0.1608242987 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_137338_010 |
0.1705520313 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 12 |
Hb_043127_010 |
0.1945429792 |
- |
- |
PREDICTED: uncharacterized protein LOC105648822 [Jatropha curcas] |
| 13 |
Hb_003589_100 |
0.1971782375 |
- |
- |
hypothetical protein JCGZ_03338 [Jatropha curcas] |
| 14 |
Hb_012691_060 |
0.1980780709 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like isoform X1 [Prunus mume] |
| 15 |
Hb_156850_050 |
0.2029689984 |
- |
- |
PREDICTED: uncharacterized protein LOC105635250 [Jatropha curcas] |
| 16 |
Hb_001498_050 |
0.2077657677 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 17 |
Hb_030949_010 |
0.2096293359 |
- |
- |
hypothetical protein POPTR_0001s24140g [Populus trichocarpa] |
| 18 |
Hb_006501_140 |
0.2136784637 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 19 |
Hb_000974_030 |
0.2239699515 |
- |
- |
PREDICTED: uncharacterized protein LOC105128463 isoform X1 [Populus euphratica] |
| 20 |
Hb_000752_040 |
0.2244688859 |
- |
- |
PREDICTED: uncharacterized protein LOC104884990 [Beta vulgaris subsp. vulgaris] |