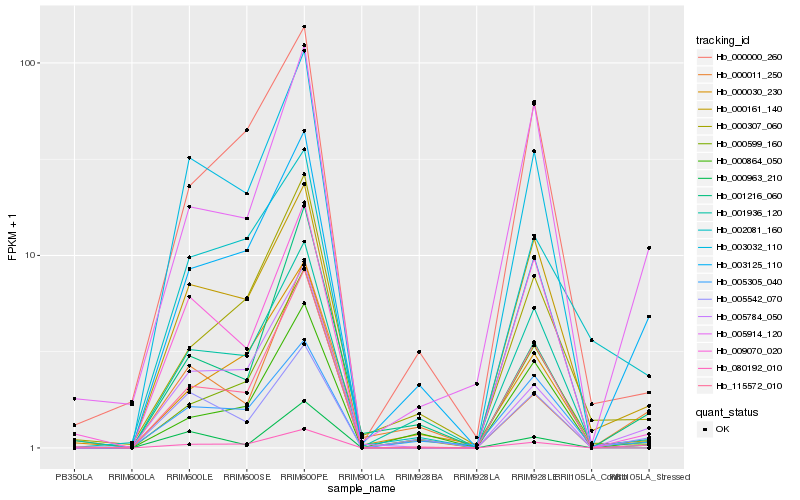
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080192_010 |
0.0 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 2 |
Hb_003125_110 |
0.1122640366 |
- |
- |
PREDICTED: protein indeterminate-domain 16 [Jatropha curcas] |
| 3 |
Hb_005784_050 |
0.1542997326 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_005914_120 |
0.1610229574 |
- |
- |
PREDICTED: actin-related protein 2/3 complex subunit 2B [Jatropha curcas] |
| 5 |
Hb_000161_140 |
0.1785282301 |
- |
- |
PREDICTED: WEB family protein At3g51220 [Jatropha curcas] |
| 6 |
Hb_005542_070 |
0.1893831025 |
- |
- |
hypothetical protein ZEAMMB73_851878 [Zea mays] |
| 7 |
Hb_000963_210 |
0.1955032891 |
- |
- |
structural constituent of cell wall, putative [Ricinus communis] |
| 8 |
Hb_001216_060 |
0.199349491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002081_160 |
0.2006581453 |
- |
- |
hypothetical protein OsI_35059 [Oryza sativa Indica Group] |
| 10 |
Hb_000307_060 |
0.2040157917 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 11 |
Hb_000030_230 |
0.2055512711 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 12 |
Hb_000011_250 |
0.2066887014 |
- |
- |
PREDICTED: RING-H2 finger protein ATL3-like [Jatropha curcas] |
| 13 |
Hb_003032_110 |
0.2077040445 |
- |
- |
- |
| 14 |
Hb_005305_040 |
0.2078144997 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 15 |
Hb_000599_160 |
0.2079879455 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 16 |
Hb_000864_050 |
0.2098436991 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_001936_120 |
0.2105211323 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 18 |
Hb_115572_010 |
0.2106974294 |
- |
- |
PREDICTED: beta-galactosidase 6 [Jatropha curcas] |
| 19 |
Hb_000000_260 |
0.2114760596 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 20 |
Hb_009070_020 |
0.2120083909 |
- |
- |
PREDICTED: uncharacterized protein LOC102612959 [Citrus sinensis] |