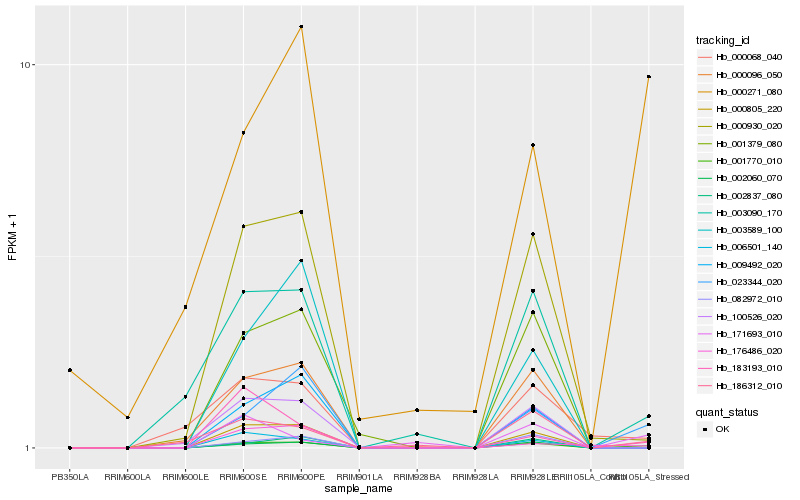
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001770_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_15779 [Jatropha curcas] |
| 2 |
Hb_023344_020 |
0.1752151138 |
- |
- |
- |
| 3 |
Hb_000930_020 |
0.2400202066 |
- |
- |
hypothetical protein PRUPE_ppa026493mg [Prunus persica] |
| 4 |
Hb_003090_170 |
0.2434704917 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 5 |
Hb_186312_010 |
0.250935709 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Populus euphratica] |
| 6 |
Hb_000805_220 |
0.2588668905 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_002060_070 |
0.2699988508 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 8 |
Hb_009492_020 |
0.2778486935 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 9 |
Hb_000096_050 |
0.2795606629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001379_080 |
0.2814498592 |
- |
- |
PREDICTED: WAT1-related protein At5g07050-like [Vitis vinifera] |
| 11 |
Hb_006501_140 |
0.283811684 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 12 |
Hb_100526_020 |
0.2848546491 |
- |
- |
hypothetical protein POPTR_0003s09340g [Populus trichocarpa] |
| 13 |
Hb_000271_080 |
0.2850370308 |
- |
- |
hypothetical protein POPTR_0006s08460g [Populus trichocarpa] |
| 14 |
Hb_000068_040 |
0.2886511515 |
- |
- |
Inter-alpha-trypsin inhibitor heavy chain H3 precursor, putative [Ricinus communis] |
| 15 |
Hb_171693_010 |
0.291985154 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 16 |
Hb_082972_010 |
0.2937610442 |
- |
- |
- |
| 17 |
Hb_002837_080 |
0.2976216777 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 18 |
Hb_003589_100 |
0.2980596912 |
- |
- |
hypothetical protein JCGZ_03338 [Jatropha curcas] |
| 19 |
Hb_183193_010 |
0.2985378169 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 20 |
Hb_176486_020 |
0.2999640196 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Populus euphratica] |