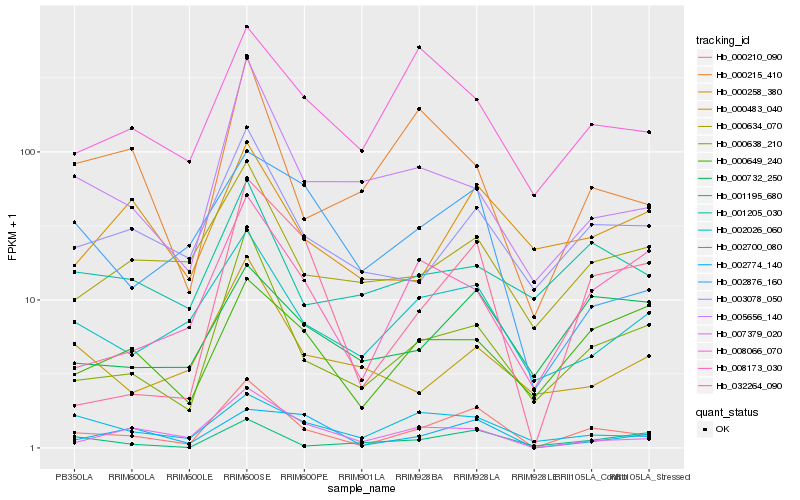
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000210_090 |
0.0 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 2 |
Hb_000638_210 |
0.1972629767 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_007379_020 |
0.2247788106 |
- |
- |
hypothetical protein JCGZ_25958 [Jatropha curcas] |
| 4 |
Hb_003078_050 |
0.2276263163 |
- |
- |
PREDICTED: serine/threonine-protein kinase OXI1 [Jatropha curcas] |
| 5 |
Hb_032264_090 |
0.2308064298 |
- |
- |
hypothetical protein JCGZ_02512 [Jatropha curcas] |
| 6 |
Hb_002026_060 |
0.2314547199 |
- |
- |
PREDICTED: uncharacterized protein LOC105645786 [Jatropha curcas] |
| 7 |
Hb_005656_140 |
0.2455461602 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 8 |
Hb_000634_070 |
0.2488819097 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g47070 [Jatropha curcas] |
| 9 |
Hb_002700_080 |
0.2499847389 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_24521 [Jatropha curcas] |
| 10 |
Hb_002774_140 |
0.2502467274 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002876_160 |
0.2555597565 |
- |
- |
sucrose transporter 1 [Hevea brasiliensis] |
| 12 |
Hb_000215_410 |
0.2571520193 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 20 [Jatropha curcas] |
| 13 |
Hb_008173_030 |
0.2579555979 |
- |
- |
PREDICTED: glutathione S-transferase T1-like [Jatropha curcas] |
| 14 |
Hb_001195_680 |
0.258746492 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 4 [Jatropha curcas] |
| 15 |
Hb_000732_250 |
0.2664502263 |
- |
- |
PREDICTED: external alternative NAD(P)H-ubiquinone oxidoreductase B2, mitochondrial-like [Populus euphratica] |
| 16 |
Hb_008066_070 |
0.2672073335 |
transcription factor |
TF Family: ERF |
AP2/ERF super family protein [Hevea brasiliensis] |
| 17 |
Hb_001205_030 |
0.2676066914 |
transcription factor |
TF Family: zf-HD |
hypothetical protein POPTR_0007s12970g [Populus trichocarpa] |
| 18 |
Hb_000649_240 |
0.2709770925 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 19 |
Hb_000483_040 |
0.2711512429 |
- |
- |
Cell division protein ftsH, putative [Ricinus communis] |
| 20 |
Hb_000258_380 |
0.2711930031 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP11-like [Jatropha curcas] |