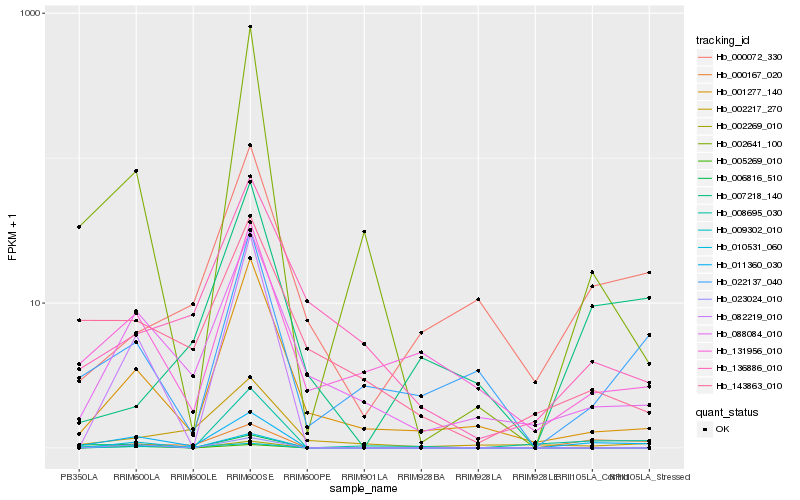
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011360_030 |
0.0 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 2 |
Hb_000167_020 |
0.2438424588 |
- |
- |
- |
| 3 |
Hb_088084_010 |
0.2671750398 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 4 |
Hb_001277_140 |
0.2879286217 |
- |
- |
PREDICTED: plasma membrane calcium-transporting ATPase 2-like [Takifugu rubripes] |
| 5 |
Hb_002641_100 |
0.2880266914 |
- |
- |
PREDICTED: uncharacterized protein LOC105115446 [Populus euphratica] |
| 6 |
Hb_022137_040 |
0.3008719694 |
- |
- |
PREDICTED: MATE efflux family protein LAL5-like [Jatropha curcas] |
| 7 |
Hb_131956_010 |
0.3121201863 |
- |
- |
hypothetical protein JCGZ_19531 [Jatropha curcas] |
| 8 |
Hb_008695_030 |
0.3149840903 |
- |
- |
hypothetical protein JCGZ_12043 [Jatropha curcas] |
| 9 |
Hb_023024_010 |
0.3166107248 |
- |
- |
PREDICTED: uncharacterized protein LOC105767094 [Gossypium raimondii] |
| 10 |
Hb_082219_010 |
0.3210006227 |
- |
- |
Os03g0264300, partial [Oryza sativa Japonica Group] |
| 11 |
Hb_143863_010 |
0.321918134 |
- |
- |
PREDICTED: uncharacterized protein LOC105113187 isoform X2 [Populus euphratica] |
| 12 |
Hb_010531_060 |
0.3224223539 |
- |
- |
hypothetical protein B456_010G209400, partial [Gossypium raimondii] |
| 13 |
Hb_000072_330 |
0.3325957601 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_006816_510 |
0.3327545799 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 15 |
Hb_007218_140 |
0.3339854202 |
- |
- |
hypothetical protein JCGZ_24847 [Jatropha curcas] |
| 16 |
Hb_005269_010 |
0.3363198093 |
- |
- |
- |
| 17 |
Hb_009302_010 |
0.3404180737 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 18 |
Hb_002217_270 |
0.3446050438 |
- |
- |
PREDICTED: desiccation-related protein PCC13-62-like [Jatropha curcas] |
| 19 |
Hb_136886_010 |
0.3454525496 |
- |
- |
hypothetical protein POPTR_0013s02830g [Populus trichocarpa] |
| 20 |
Hb_002269_010 |
0.3474182455 |
- |
- |
PREDICTED: WAT1-related protein At4g15540-like [Jatropha curcas] |