| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
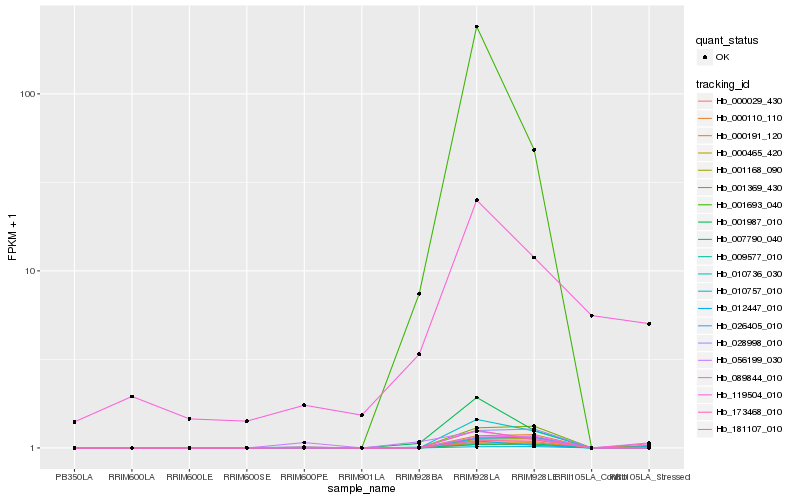
Hb_173468_010 |
0.0 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 2 |
Hb_000465_420 |
0.210865535 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 3 |
Hb_000191_120 |
0.222949269 |
- |
- |
- |
| 4 |
Hb_012447_010 |
0.2254150209 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_007790_040 |
0.2827444007 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 6 |
Hb_010736_030 |
0.2861283989 |
- |
- |
- |
| 7 |
Hb_010757_010 |
0.306333 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 8 |
Hb_056199_030 |
0.32723422 |
- |
- |
PREDICTED: purine permease 3-like isoform X2 [Gossypium raimondii] |
| 9 |
Hb_119504_010 |
0.3279657937 |
- |
- |
hypothetical protein JCGZ_15304 [Jatropha curcas] |
| 10 |
Hb_001693_040 |
0.3299643479 |
- |
- |
- |
| 11 |
Hb_001987_010 |
0.3308493412 |
- |
- |
- |
| 12 |
Hb_001369_430 |
0.3344025578 |
- |
- |
PREDICTED: uncharacterized protein LOC105648869 [Jatropha curcas] |
| 13 |
Hb_028998_010 |
0.3347185169 |
- |
- |
- |
| 14 |
Hb_000110_110 |
0.3353156361 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 15 |
Hb_089844_010 |
0.3354622125 |
- |
- |
- |
| 16 |
Hb_026405_010 |
0.3355683045 |
- |
- |
PREDICTED: uncharacterized protein LOC104107896 [Nicotiana tomentosiformis] |
| 17 |
Hb_009577_010 |
0.3355914761 |
- |
- |
- |
| 18 |
Hb_001168_090 |
0.3357046927 |
- |
- |
PREDICTED: uncharacterized protein LOC104246050 [Nicotiana sylvestris] |
| 19 |
Hb_181107_010 |
0.335853814 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 20 |
Hb_000029_430 |
0.3360048775 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic-like [Jatropha curcas] |