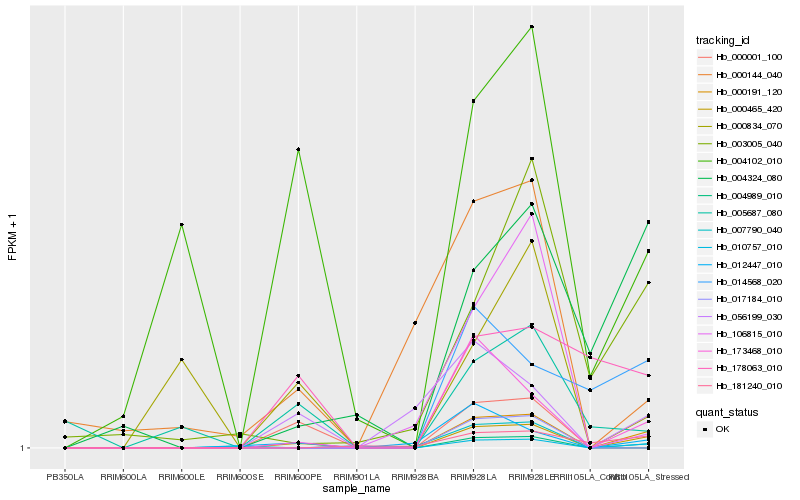
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000465_420 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 2 |
Hb_000191_120 |
0.1973250577 |
- |
- |
- |
| 3 |
Hb_173468_010 |
0.210865535 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_010757_010 |
0.2893874893 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 5 |
Hb_012447_010 |
0.3009037078 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_007790_040 |
0.3019722678 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 7 |
Hb_056199_030 |
0.3120328642 |
- |
- |
PREDICTED: purine permease 3-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_004324_080 |
0.3377173482 |
- |
- |
- |
| 9 |
Hb_000001_100 |
0.3428875722 |
- |
- |
- |
| 10 |
Hb_178063_010 |
0.3495914062 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 11 |
Hb_106815_010 |
0.3519926634 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 12 |
Hb_181240_010 |
0.3531187012 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 13 |
Hb_003005_040 |
0.3588371583 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000834_070 |
0.3608868408 |
- |
- |
PREDICTED: tankyrase-1 isoform X4 [Jatropha curcas] |
| 15 |
Hb_014568_020 |
0.3635869519 |
- |
- |
PREDICTED: GPI mannosyltransferase 2 [Jatropha curcas] |
| 16 |
Hb_004102_010 |
0.3641479212 |
- |
- |
F-box family protein [Theobroma cacao] |
| 17 |
Hb_005687_080 |
0.372013569 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 18 |
Hb_000144_040 |
0.3751130044 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 19 |
Hb_017184_010 |
0.3806894353 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 20 |
Hb_004989_010 |
0.3806894937 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |