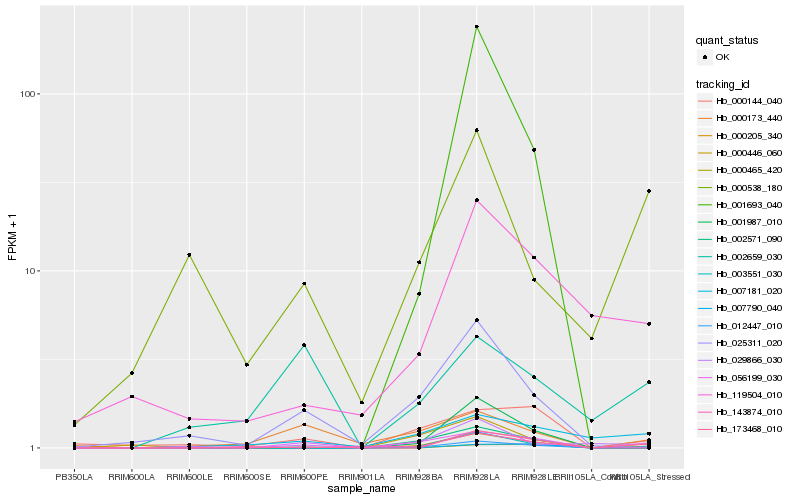
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012447_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_056199_030 |
0.2076548892 |
- |
- |
PREDICTED: purine permease 3-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_173468_010 |
0.2254150209 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 4 |
Hb_007790_040 |
0.2791856702 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 5 |
Hb_007181_020 |
0.2921250401 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Populus euphratica] |
| 6 |
Hb_119504_010 |
0.2939597425 |
- |
- |
hypothetical protein JCGZ_15304 [Jatropha curcas] |
| 7 |
Hb_000465_420 |
0.3009037078 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 8 |
Hb_000144_040 |
0.3014897368 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 9 |
Hb_025311_020 |
0.3087711819 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 10 |
Hb_000173_440 |
0.3144840989 |
- |
- |
YALI0B05280p [Yarrowia lipolytica] |
| 11 |
Hb_000205_340 |
0.3148882386 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 12 |
Hb_001987_010 |
0.3187273969 |
- |
- |
- |
| 13 |
Hb_029866_030 |
0.3215013572 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |
| 14 |
Hb_003551_030 |
0.3235477405 |
- |
- |
hypothetical protein POPTR_0016s03850g [Populus trichocarpa] |
| 15 |
Hb_000446_060 |
0.3255942025 |
- |
- |
- |
| 16 |
Hb_000538_180 |
0.3401203503 |
- |
- |
PREDICTED: probable polyamine oxidase 4 [Jatropha curcas] |
| 17 |
Hb_002571_090 |
0.3408776447 |
- |
- |
- |
| 18 |
Hb_143874_010 |
0.3409790601 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 19 |
Hb_001693_040 |
0.3421163667 |
- |
- |
- |
| 20 |
Hb_002659_030 |
0.3427335353 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |