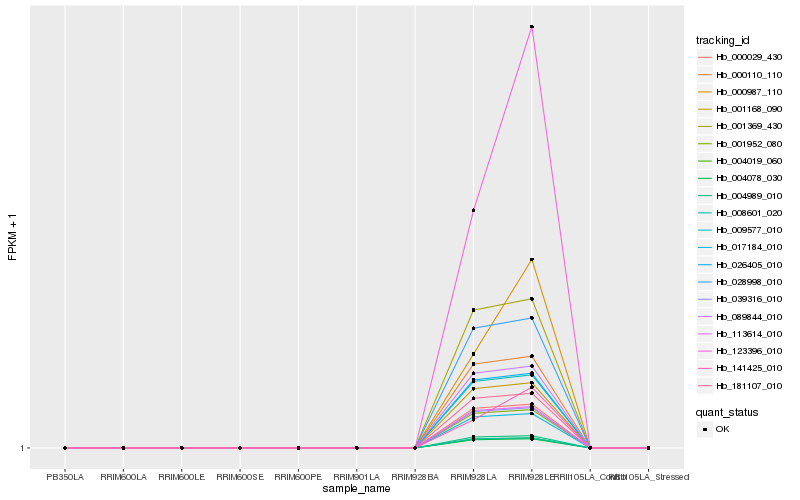
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017184_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 2 |
Hb_001952_080 |
0.0001053252 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 3 |
Hb_039316_010 |
0.0001573554 |
- |
- |
PREDICTED: uncharacterized protein LOC105771917 [Gossypium raimondii] |
| 4 |
Hb_113614_010 |
0.000186452 |
- |
- |
hypothetical protein VITISV_001308 [Vitis vinifera] |
| 5 |
Hb_000029_430 |
0.0002517315 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_181107_010 |
0.0005489156 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 7 |
Hb_004989_010 |
0.0005732484 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 8 |
Hb_008601_020 |
0.0006243936 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_004019_060 |
0.000630521 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 10 |
Hb_004078_030 |
0.0006520038 |
- |
- |
Putative polyprotein, identical [Solanum demissum] |
| 11 |
Hb_001168_090 |
0.0008425928 |
- |
- |
PREDICTED: uncharacterized protein LOC104246050 [Nicotiana sylvestris] |
| 12 |
Hb_009577_010 |
0.0010657689 |
- |
- |
- |
| 13 |
Hb_026405_010 |
0.0011114679 |
- |
- |
PREDICTED: uncharacterized protein LOC104107896 [Nicotiana tomentosiformis] |
| 14 |
Hb_089844_010 |
0.001320799 |
- |
- |
- |
| 15 |
Hb_000110_110 |
0.0016102735 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 16 |
Hb_028998_010 |
0.0027927092 |
- |
- |
- |
| 17 |
Hb_001369_430 |
0.0034204726 |
- |
- |
PREDICTED: uncharacterized protein LOC105648869 [Jatropha curcas] |
| 18 |
Hb_123396_010 |
0.1357971958 |
- |
- |
hypothetical protein AMTR_s00510p00011440 [Amborella trichopoda] |
| 19 |
Hb_000987_110 |
0.1406837469 |
- |
- |
PREDICTED: uncharacterized protein LOC104902780 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_141425_010 |
0.1425414005 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |