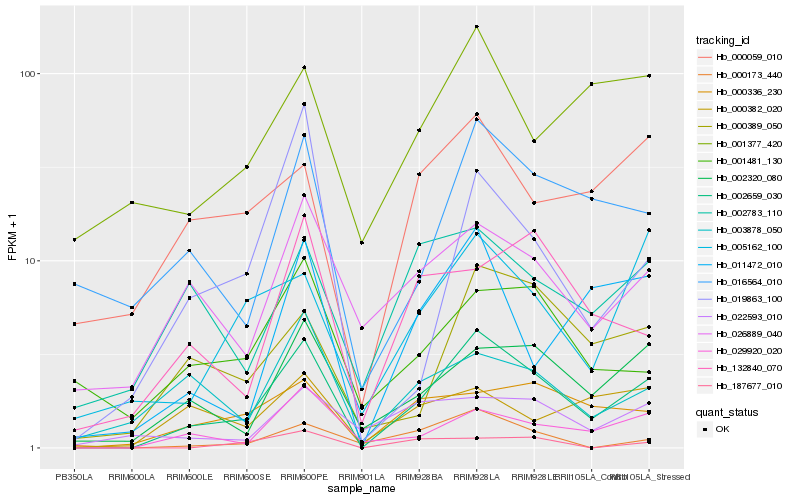
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002659_030 |
0.0 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 2 |
Hb_029920_020 |
0.2086502477 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG7 [Jatropha curcas] |
| 3 |
Hb_002320_080 |
0.2106566133 |
- |
- |
PREDICTED: MLO-like protein 13 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000389_050 |
0.2329721262 |
- |
- |
PREDICTED: uncharacterized protein LOC105650917 [Jatropha curcas] |
| 5 |
Hb_016564_010 |
0.2351027849 |
- |
- |
PREDICTED: uncharacterized protein LOC101764501 [Setaria italica] |
| 6 |
Hb_000336_230 |
0.2360724262 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa007609mg [Prunus persica] |
| 7 |
Hb_011472_010 |
0.2371275651 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 8 |
Hb_026889_040 |
0.2427435776 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 9 |
Hb_005162_100 |
0.2462908618 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_003878_050 |
0.2503737317 |
- |
- |
PREDICTED: peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A-like [Jatropha curcas] |
| 11 |
Hb_000059_010 |
0.2537070567 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_002783_110 |
0.2560699547 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 13 |
Hb_000173_440 |
0.2564375811 |
- |
- |
YALI0B05280p [Yarrowia lipolytica] |
| 14 |
Hb_132840_070 |
0.2575192193 |
- |
- |
metal tolerance protein 1 [Populus trichocarpa x Populus deltoides] |
| 15 |
Hb_001481_130 |
0.2610225572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_022593_010 |
0.264910027 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 17 |
Hb_019863_100 |
0.2663412719 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001377_420 |
0.2676659183 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000382_020 |
0.2698770533 |
- |
- |
PREDICTED: uncharacterized protein LOC105633610 [Jatropha curcas] |
| 20 |
Hb_187677_010 |
0.2703840589 |
- |
- |
- |